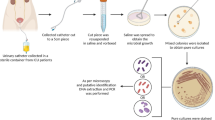
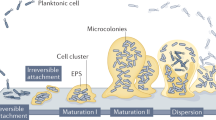
Abstract
Biofilms are structured microbial communities embedded within an extracellular matrix that confers protection against environmental stresses. In both natural and clinical settings, biofilms are rarely composed of a single species and may also involve interactions with bacteriophages or even eukaryotic viruses. Since both biofilms and viruses are ubiquitous, and viruses remain among the neglected components of the microbiome, understanding their interactions is essential. In hospitalized patients, catheter colonization by biofilms markedly increases the risk of bacteremia and septicemia, and biofilm formation is almost inevitable during long-term catheterization. In this study, we investigated biofilm-forming capacities of uropathogenic Escherichia coli (UPEC) and clinical strains associated with catheter-related systemic infections. Selected strains were further examined to evaluate the influence of the ubiquitous mammalian reovirus on bacterial biofilm formation and to evaluate biofilm entrapment of viral particles and its impact on viral infectivity. Bacterial growth, survival and biofilm production were measured in the presence or absence of the virus. While reovirus exhibited no bactericidal effects and biofilm biomass remained largely unchanged, rheological and microscopic analyses revealed strain-specific alterations in biofilm properties. Remarkably, reovirus retains infectivity after release from biofilms, indicating that bacterial biofilms may serve as reservoirs or shelters for eukaryotic viruses.
Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on request.
References
Flemming, H. C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. https://doi.org/10.1038/nrmicro.2016.94 (2016).
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. https://doi.org/10.1126/science.284.5418.1318 (1999).
Hoyle, B. D. & Costerton, J. W. Bacterial resistance to antibiotics: the role of biofilms. Prog Drug Res. 37, 91–105. https://doi.org/10.1007/978-3-0348-7139-6_2 (1991).
Bryers, J. D. Medical biofilms. Biotechnol. Bioeng. 100, 1–18. https://doi.org/10.1002/bit.21838 (2008).
Trautner, B. W. & Darouiche, R. O. Role of biofilm in catheter-associated urinary tract infection. Am. J. Infect. Control. 32, 177–183. https://doi.org/10.1016/j.ajic.2003.08.005 (2004).
Gahlot, R., Nigam, C., Kumar, V., Yadav, G. & Anupurba, S. Catheter-related bloodstream infections. Int. J. Crit. Illn. Inj Sci. 4, 162–167. https://doi.org/10.4103/2229-5151.134184 (2014).
Azevedo, A. S., Almeida, C., Melo, L. F. & Azevedo, N. F. Impact of polymicrobial biofilms in catheter-associated urinary tract infections. Crit. Rev. Microbiol. 43, 423–439. https://doi.org/10.1080/1040841X.2016.1240656 (2017).
Wilks, S. A., Koerfer, V. V., Prieto, J. A., Fader, M. & Keevil, C. W. Biofilm Development on Urinary Catheters Promotes the Appearance of Viable but Nonculturable Bacteria. mBio 12, (2021). https://doi.org/10.1128/mBio.03584-20
Donlan, R. M. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. https://doi.org/10.3201/eid0809.020063 (2002).
Flores-Mireles, A., Hreha, T. N., Hunstad, D. A. & Pathophysiology Treatment, and prevention of Catheter-Associated urinary tract infection. Top. Spinal Cord Inj Rehabil. 25, 228–240. https://doi.org/10.1310/sci2503-228 (2019).
Peters, B. M., Jabra-Rizk, M. A., O’May, G. A., Costerton, J. W. & Shirtliff, M. E. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25, 193–213. https://doi.org/10.1128/CMR.00013-11 (2012).
Sadiq, F. A. et al. Trans-kingdom interactions in mixed biofilm communities. FEMS Microbiol. Rev. 46 https://doi.org/10.1093/femsre/fuac024 (2022).
Elias, S. & Banin, E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004. https://doi.org/10.1111/j.1574-6976.2012.00325.x (2012).
Brogden, K. A., Guthmiller, J. M. & Taylor, C. E. Human polymicrobial infections. Lancet 365, 253–255. https://doi.org/10.1016/S0140-6736(05)17745-9 (2005).
Kilic, T. in Exploring bacterial biofilms (ed InTech)InTech, (2025).
Ugwu, C. et al. Biofilms: structure, resistance mechanism, emerging control strategies, and applications. RCS Pharm. 6 https://doi.org/10.1039/D5PM00094G (2025).
Goller, C. C. & Romeo, T. Environmental influences on biofilm development. Curr. Top. Microbiol. Immunol. 322, 37–66. https://doi.org/10.1007/978-3-540-75418-3_3 (2008).
Biswas, T., Ahmed, M. & Mondal, S. Mixed species biofilm: Structure, challenge and its intricate involvement in hospital associated infections. Microb. Pathog. 195, 106866. https://doi.org/10.1016/j.micpath.2024.106866 (2024).
Joshi, R. V., Gunawan, C. & Mann, R. We are one: multispecies metabolism of a biofilm consortium and their treatment strategies. Front. Microbiol. 12, 635432. https://doi.org/10.3389/fmicb.2021.635432 (2021).
Gordon, V., Bakhtiari, L. & Kovach, K. From molecules to multispecies ecosystems: the roles of structure in bacterial biofilms. Phys. Biol. 16, 041001. https://doi.org/10.1088/1478-3975/ab1384 (2019).
Visnapuu, A., Van der Gucht, M., Wagemans, J. & Lavigne, R. Deconstructing the Phage-Bacterial biofilm interaction as a basis to Establish new antibiofilm strategies. Viruses 14 https://doi.org/10.3390/v14051057 (2022).
Sutherland, I. W., Hughes, K. A., Skillman, L. C. & Tait, K. The interaction of phage and biofilms. FEMS Microbiol. Lett. 232, 1–6. https://doi.org/10.1016/S0378-1097(04)00041-2 (2004).
Chang, C. et al. Bacteriophage-Mediated control of biofilm: A promising new dawn for the future. Front. Microbiol. 13, 825828. https://doi.org/10.3389/fmicb.2022.825828 (2022).
Burkle, M. et al. Phage-phage competition and biofilms affect interactions between two virulent bacteriophages and Pseudomonas aeruginosa. ISME J. 19 https://doi.org/10.1093/ismejo/wraf065 (2025).
Wingender, J. & Flemming, H. C. Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Environ. Health. 214, 417–423. https://doi.org/10.1016/j.ijheh.2011.05.009 (2011).
Quignon, F., Sardin, M., Kiene, L. & Schwartzbrod, L. Poliovirus-1 inactivation and interaction with biofilm: a pilot-scale study. Appl. Environ. Microbiol. 63, 978–982. https://doi.org/10.1128/aem.63.3.978-982.1997 (1997).
Ascione, C. et al. Herpes simplex virus-1 entrapped in Candida albicans biofilm displays decreased sensitivity to antivirals and UVA1 laser treatment. Ann. Clin. Microbiol. Antimicrob. 16 https://doi.org/10.1186/s12941-017-0246-5 (2017).
Lacroix-Gueu, P., Briandet, R., Leveque-Fort, S. & Bellon-Fontaine, M. N. Fontaine-Aupart, M. P. In situ measurements of viral particles diffusion inside mucoid biofilms. C R Biol. 328, 1065–1072. https://doi.org/10.1016/j.crvi.2005.09.010 (2005).
Wojciuk, B. et al. Urobiome. Sickness Health Microorganisms. 7 https://doi.org/10.3390/microorganisms7110548 (2019).
Whiteside, S. A., Razvi, H., Dave, S., Reid, G. & Burton, J. P. The Microbiome of the urinary tract–a role beyond infection. Nat. Rev. Urol. 12, 81–90. https://doi.org/10.1038/nrurol.2014.361 (2015).
Bjerre, R. D. et al. Skin dysbiosis in the Microbiome in atopic dermatitis is site-specific and involves bacteria, fungus and virus. BMC Microbiol. 21, 256. https://doi.org/10.1186/s12866-021-02302-2 (2021).
Chen, Y. E. & Tsao, H. The skin microbiome: current perspectives and future challenges. J. Am. Acad. Dermatol. 69, 143–155. https://doi.org/10.1016/j.jaad.2013.01.016 (2013).
Berger, A. K., Yi, H., Kearns, D. B. & Mainou, B. A. Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog. 13, e1006768. https://doi.org/10.1371/journal.ppat.1006768 (2017).
Hollenbeck, E. C. et al. Molecular determinants of mechanical properties of V. cholerae biofilms at the air-liquid interface. Biophys. J. 107, 2245–2252. https://doi.org/10.1016/j.bpj.2014.10.015 (2014).
Abriat, C. et al. Mechanical and microstructural insights of Vibrio cholerae and Escherichia coli dual-species biofilm at the air-liquid interface. Colloids Surf. B Biointerfaces. 188, 110786. https://doi.org/10.1016/j.colsurfb.2020.110786 (2020).
Abriat, C., Virgilio, N., Heuzey, M. C. & Daigle, F. Microbiological and real-time mechanical analysis of Bacillus licheniformis and Pseudomonas fluorescens dual-species biofilm. Microbiol. (Reading). 165, 747–756. https://doi.org/10.1099/mic.0.000819 (2019).
Charlton, S. G. V. et al. Regulating, Measuring, and modeling the viscoelasticity of bacterial biofilms. J. Bacteriol. 201 https://doi.org/10.1128/JB.00101-19 (2019).
Beckwith, J., Ganesan, K., VanEpps, M., Kumar, J. S., Solomon, M. & A. & J. Rheology of Candida albicans fungal biofilms. J. Rheol. 66, 683–697 (2022).
Serra, D. O., Richter, A. M. & Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 195, 5540–5554. https://doi.org/10.1128/JB.00946-13 (2013).
Mobley, H. L. et al. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58, 1281–1289. https://doi.org/10.1128/iai.58.5.1281-1289.1990 (1990).
Kanai, Y., Kawagishi, T., Matsuura, Y. & Kobayashi, T. In vivo live imaging of oncolytic mammalian orthoreovirus expressing nanoluc luciferase in tumor xenograft mice. J. Virol. 93 https://doi.org/10.1128/JVI.00401-19 (2019).
Hassanbhai, A. M., Phoon, M. C., Chow, V. T. & Ho, B. The association of Helicobacter pylori biofilm with enterovirus 71 prolongs viral viability and survival. Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms241914500 (2023).
Greaves, J. C. et al. Extended persistence and thermostability of adenovirus 41 in the presence of bacteria. J. Appl. Microbiol. https://doi.org/10.1093/jambio/lxaf221 (2025).
Gagné, M. J., Savard, T. & Brassard, J. Interactions between infectious foodborne viruses and bacterial biofilms formed on different food contact surfaces. Food Environ. Virol. 14, 267–279. https://doi.org/10.1007/s12560-022-09534-z (2022).
Hendricks, M. R. et al. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl. Acad. Sci. U S A. 113, 1642–1647. https://doi.org/10.1073/pnas.1516979113 (2016).
Kiedrowski, M. R. et al. Staphylococcus aureus Biofilm Growth on Cystic Fibrosis Airway Epithelial Cells Is Enhanced during Respiratory Syncytial Virus Coinfection. mSphere 3, (2018). https://doi.org/10.1128/mSphere.00341-18
Hall, J. et al. Effects of influenza A infection on liberation of bacteria from biofilms and inflammatory response in an in vitro model of chronic rhinosinusitis. Microbiol. (Reading). 171. https://doi.org/10.1099/mic.0.001586 (2025).
Johnson, J. R., Moseley, S. L., Roberts, P. L. & Stamm, W. E. Aerobactin and other virulence factor genes among strains of Escherichia coli causing urosepsis: association with patient characteristics. Infect. Immun. 56, 405–412. https://doi.org/10.1128/iai.56.2.405-412.1988 (1988).
Johnson, J. R. et al. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11, 141–145. https://doi.org/10.3201/eid1101.040418 (2005).
Manges, A. R., Tabor, H., Tellis, P., Vincent, C. & Tellier, P. P. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg. Infect. Dis. 14, 1575–1583. https://doi.org/10.3201/eid1410.080102 (2008).
Guyomard-Rabenirina, S. et al. Temporal trends and risks factors for antimicrobial resistant Enterobacteriaceae urinary isolates from outpatients in Guadeloupe. BMC Microbiol. 16, 121. https://doi.org/10.1186/s12866-016-0749-9 (2016).
Sandekian, V. & Lemay, G. A single amino acid substitution in the mRNA capping enzyme lambda2 of a mammalian orthoreovirus mutant increases interferon sensitivity. Virology 483, 229–235. https://doi.org/10.1016/j.virol.2015.04.020 (2015).
Danis, C. & Lemay, G. Protein synthesis in different cell lines infected with orthoreovirus serotype 3: Inhibition of host-cell protein synthesis correlates with accelerated viral multiplication and cell killing. Biochem. Cell. Biol. 71, 81–85. https://doi.org/10.1139/o93-012 (1993).
Coombs, K. M. & Mammalian Reoviruses Propagation, Quantification, and storage. Curr. Protoc. 3, e716. https://doi.org/10.1002/cpz1.716 (2023).
Georgi, A., Mottola-Hartshorn, C., Warner, A., Fields, B. & Chen, L. B. Detection of individual fluorescently labeled reovirions in living cells. Proc. Natl. Acad. Sci. U S A. 87, 6579–6583. https://doi.org/10.1073/pnas.87.17.6579 (1990).
Lanoie, D. & Lemay, G. Multiple proteins differing between laboratory stocks of mammalian orthoreoviruses affect both virus sensitivity to interferon and induction of interferon production during infection. Virus Res. 247, 40–46. https://doi.org/10.1016/j.virusres.2018.01.009 (2018).
Desloges, I. et al. Identification and characterization of OmpT-like proteases in uropathogenic Escherichia coli clinical isolates. Microbiologyopen 8, e915. https://doi.org/10.1002/mbo3.915 (2019).
Johnson, J. R. et al. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, Iha and iron (E. coli), among Escherichia coli isolates from patients with Urosepsis. Infect. Immun. 68, 3040–3047. https://doi.org/10.1128/IAI.68.5.3040-3047.2000 (2000).
Funding
This work was supported by a grant NFRFE-2022-00464 from the New Frontiers in Research Fund and from the Swine and Poultry Infectious Diseases Research Center (CRIPA) funded by Fonds de recherche du Québec - Nature et technologies (FRQNT).
Author information
Authors and Affiliations
Contributions
JG performed the bacterial growth, survival and biofilms experiments. CA conducted the rheological analyses. MLH produced the virus stock and luminescent constructs. GL and MCH contributed to data analysis and interpretation. CMD and CQ provided bacterial strains. NV, GL, CQ, MCH and FD secured funding. FD was the major contributor to manuscript writing. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, J., Abriat, C., Laekas-Hameder, M. et al. Microbiological and rheological dynamics of mixed biofilms formed by bacteria and eukaryotic virus. Sci Rep (2026). https://doi.org/10.1038/s41598-026-39314-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-39314-9