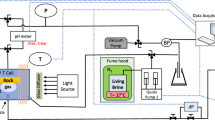
Abstract
Bioleaching is a prominent method among eco-friendly techniques. The efficiency of bioleaching is reduced by the presence of brackish water. Moreover, it has been reported that the most successful bioleaching systems are those that include both autotrophic and heterotrophic microorganismes. This research focuses on enhancing uranium bioleaching in brackish waters using the halotolerant bacterium Acidithiobacillus ferrooxidans strain THA4 and the fungus Rhodotorula toruloides strain IR-1395. The experimental data were modeled by the Response Surface Methodology (RSM) approach. The suggested model for uranium extraction in a brackish environment using microorganisms demonstrated an alignment with the experimental data, with a correlation coefficient of R2 = 0.94. The results showed that the amount of uranium bioleaching by the consortium increased by 24.22%, compared to the bacterium alone under the optimal conditions suggested by the software. The study employed SEM–EDS to investigate the morphological changes in ore samples exposed to the microorganisms. The findings offer insights into the relationship dynamics between acidophilic bacteria and heterotrophic yeasts in uranium bioleaching of brackish waters. Finally, this study has improved biohydrometallurgical methods for uranium extraction from low-grade ores, especially in saline and low-resources conditions.
Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this article. The 16S rDNA sequence of Acidithiobacillus ferrooxidans strain THA4 had been deposited in the National Center for Biotechnology Information (NCBI) GenBank with the accession number PV981793 (https://www.ncbi.nlm.nih.gov/nuccore/PV981793). The 18S rDNA sequence and large subunit rDNA sequence of Rhodotorula toruloides strain IR-1395 had been deposited in the National Center for Biotechnology Information (NCBI) GenBank with the accession number KX452402.1 and MN752210.1, respectively (https://www.ncbi.nlm.nih.gov/nuccore/KX452402 and https://www.ncbi.nlm.nih.gov/nuccore/MN752210).
References
Tavakoli, H. Z., Abdollahy, M., Ahmadi, S. & Darban, A. K. Kinetics of uranium bioleaching in stirred and column reactors. Miner. Eng. 111, 36–46 (2017).
Kaksonen, A. H., Lakaniemi, A.-M. & Tuovinen, O. H. Acid and ferric sulfate bioleaching of uranium ores: A review. J. Clean. Prod. 264, 121586 (2020).
Pradhan, N., Nathsarma, K., Rao, K. S., Sukla, L. & Mishra, B. Heap bioleaching of chalcopyrite: A review. Miner. Eng. 21(5), 355–365 (2008).
Chen, W., Tang, H. & Yin, S. Bioleaching of low-grade copper sulfide enhanced by nutrients from sterilized medical waste. Process Saf. Environ. Protect. 188, 1527–1535 (2024).
Nagpal, S., Dahlstrom, D. & Oolman, T. Effect of carbon dioxide concentration on the bioleaching of a pyrite–arsenopyrite ore concentrate. Biotechnol. Bioeng. 41(4), 459–464 (1993).
Barron, J. L. & Lueking, D. R. Growth and maintenance of Thiobacillus ferrooxidans cells. Appl. Environ. Microbiol. 56(9), 2801–2806 (1990).
Schippers, A., Hetz, S. A. & Ostertag-Henning, C. Laterite ore processing with hydrogen via mild chemical pressure leaching or bioleaching. Hydrometallurgy https://doi.org/10.1016/j.hydromet.2025.106447 (2025).
Khetwunchai, N. et al. Enhanced bioleaching of copper and gold from waste printed circuit boards: Stepwise process, pretreatment strategies, metabolomics analysis, and the role of N8-acetylspermidine. Process Saf. Environ. Protect. 194, 289–305 (2025).
Dew, D. W., Lawson, F. & Broadhurst, J. L. The bioleaching of sulfide minerals with emphasis on copper sulfides a review. Hydrometallurgy 47, 155–170 (1997).
Rea, S. et al. Salt-tolerant microorganisms potentially useful for bioleaching operations where fresh water is scarce. Miner. Eng. 75, 126–132 (2015).
Simmons, S. F. & Norris, P. R. Acidophilic microorganisms and their interactions with minerals in saline environments. Extremophiles 6(6), 551–559 (2002).
Noguchi, H. & Okibe, N. The role of bioleaching microorganisms in saline water leaching of chalcopyrite concentrate. Hydrometallurgy 195, 105397 (2020).
Shivanand, P. & Mugeraya, G. Halophilic microorganisms and their adaptation mechanisms. Crit. Rev. Microbiol. 37(4), 315–334 (2011).
Oren, A. Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 4, 315 (2013).
Graziano, G. & Merlino, A. Molecular bases of protein halotolerance. Biochimica et Biophysica Acta (BBA) 1844(4), 850–858 (2014).
Fournier, D., Lemieux, R. & Couillard, D. Essential interactions between Thiobacillus ferrooxidans and heterotrophic microorganisms during a wastewater sludge bioleaching process. Environ. Pollut. 101(2), 303–309 (1998).
Gu, X.-Y. & Wong, J. W. Degradation of inhibitory substances by heterotrophic microorganisms during bioleaching of heavy metals from anaerobically digested sewage sludge. Chemosphere 69(2), 311–318 (2007).
Zheng, G., Zhou, L. & Wang, S. An acid-tolerant heterotrophic microorganism role in improving tannery sludge bioleaching conducted in successive multibatch reaction systems. Environ. Sci. Technol. 43(11), 4151–4156 (2009).
Tavakoli, H. Z., Abdollahy, M., Ahmadi, S. & Darban, A. K. Enhancing recovery of uranium column bioleaching by process optimization and kinetic modeling. Trans. Nonferrous Met. Soc. China 27(12), 2691–2703 (2017).
Bomberg, M., Mäkinen, J., Salo, M. & Kinnunen, P. High diversity in iron cycling microbial communities in acidic, iron‐rich water of the Pyhäsalmi Mine, Finland. Geofluids 2019(1), 7401304 (2019).
Piroeva, I. et al. A simple and rapid scanning electron microscope preparative technique for observation of biological samples: application on bacteria and DNA samples. Bulg. Chem. Commun 45(4), 510–515 (2013).
Csonka, L. N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53(1), 121–147 (1989).
Kieft, T. L. & Spence, S. D. Osmoregulation in Thiobacillus ferrooxidans: Stimulation of iron oxidation by proline and betaine under salt stress. Curr. Microbiol. 17, 255–258 (1988).
Guo, X. et al. Role of proline biosynthesis in Acidithiobacillus caldus under salt stress. J. Bacteriol. 195, 4421–4429 (2013).
Zammit, C. M. et al. Bioleaching in brackish waters—effect of chloride ions on the acidophile population and proteomes of model species. Appl. Microbiol. Biotechnol. 93, 319–329 (2012).
Simmons, S. & Norris, P. Acidophiles of saline water at thermal vents of Vulcano, Italy. Extremophiles 6, 201–207 (2002).
Sand, W. et al. Microbial mechanisms for metal leaching in acidic environments. Hydrometallurgy 59(3), 159 (2001).
Zheng, X. & Li, D. Interaction of Acidithiobacillus ferrooxidans, Rhizobium phaseoli and Rhodotorula sp. in bioleaching process based on Lotka-Volterra model. Electron. J. Biotechnol. 22, 90–97 (2016).
Ingledew, W. J. Thiobacillus ferrooxidans the bioenergetics of an acidophilic chemolithotroph. Biochimica et Biophysica Acta (BBA) 683(2), 89–117 (1982).
Salinas, E. et al. Removal of cadmium and lead from dilute aqueous solutions by Rhodotorula rubra. Bioresour. Technol. 72(2), 107–112 (2000).
Acknowledgements
This manuscript was a part of the PhD. thesis by M. Shoja, under the supervision of Dr. P. Mohammadi and Dr. P. Tajer-Mohammad-Ghazvini; and advi¬sory of H. Zare-Tavakoli. The authors would like to thank Alzahra University and also Nuclear Science and Technology Research Institute, Tehran, Iran for their support through this study.
Author information
Authors and Affiliations
Contributions
The manuscript was a part of the PhD. thesis by M. S. All the authors contributed to the conception and design of the study. M. S.: conducting experiments, writing (original draft preparation). Dr. P.M.: writing (review and editing), supervision. Dr. P. T-M-G.: writing (review and editing), supervision, software. Dr. H. Z-T.: adviser. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shoja, M., Mohammadi, P., Tajer-Mohammad-Ghazvini, P. et al. Improved uranium bioleaching in brackish environments via microbial consortium using RSM based modelling and optimization. Sci Rep (2026). https://doi.org/10.1038/s41598-026-39700-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-39700-3