Fig. 1: Principles of TAS-Seq.
From: TAS-Seq is a robust and sensitive amplification method for bead-based scRNA-seq
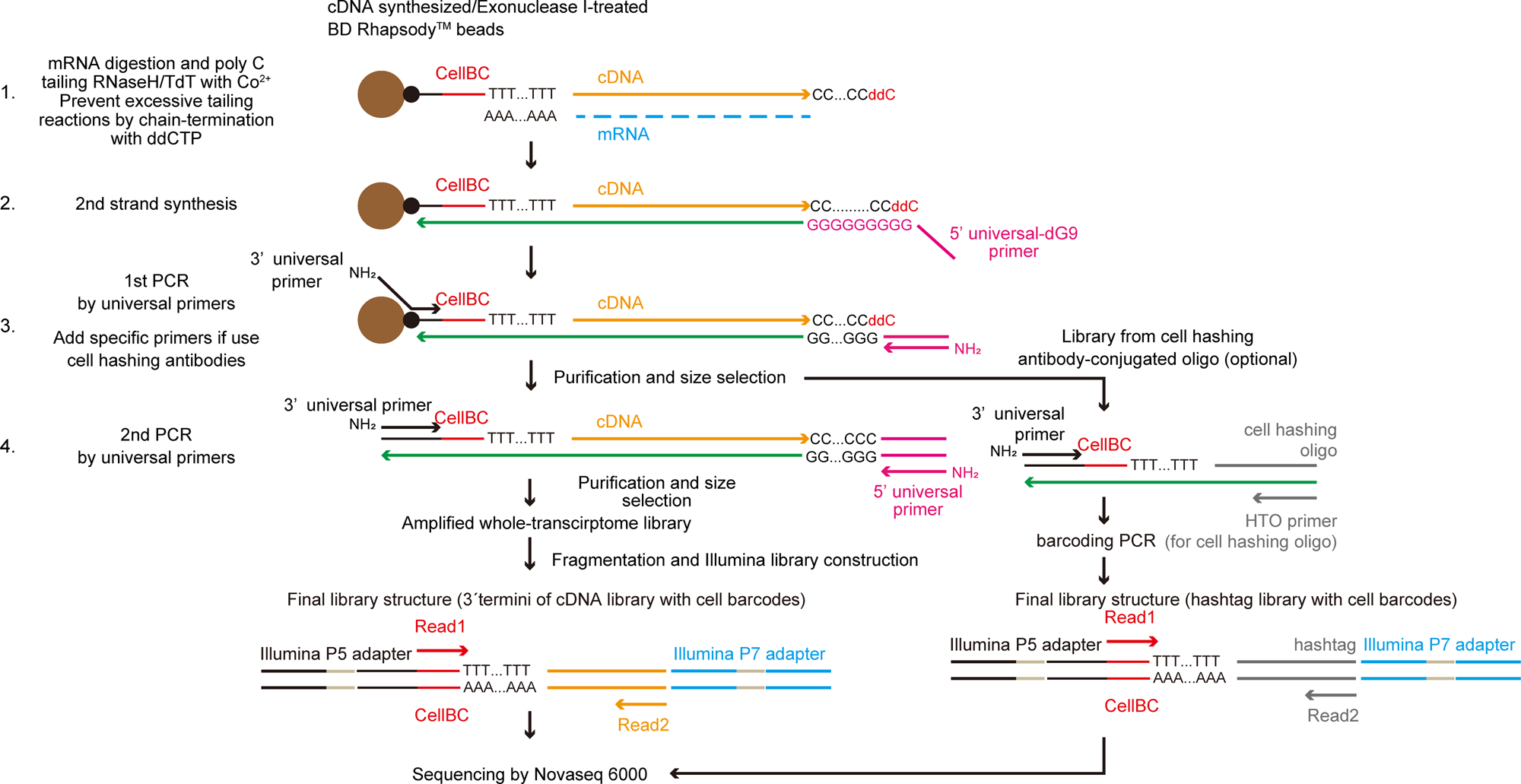
Diagram of the TAS-Seq library preparation workflow. First, cDNA synthesis on BD Rhapsody magenic beads were performed. After Exonuclease I treatment, dC-tailing reaction was performed by TdT/RNase H. Tailing reaction was stochastically stopped by ddCTP incorporation into 3' termini (1). Next, second-strand synthesis was performed using 5'universal-dG9 primer and PCR master mix (2). Then, 1st PCR was performed by appropriate primers (3′ and 5' universal primer (if only amplify cDNA) or 3′, 5' universal primer and HTO primer (if use cell hashing antibodies)) (3). Resultant libraries were size-selected and amplified by 2nd PCR (4). cDNA libraries were fragmented, ligated, and truncated with Illumina P7 adapters, and 3′ termini of cDNA molecules with cell barcodes were enriched by PCR amplification. During the PCR step, we introduced unique-dual barcodes to the Illumina adapters to minimize index hopping. Finally, libraries were sequenced by Illumina Novaseq 6000.
