Fig. 1: Selective single cell RNA sequencing using See-N-Seq.
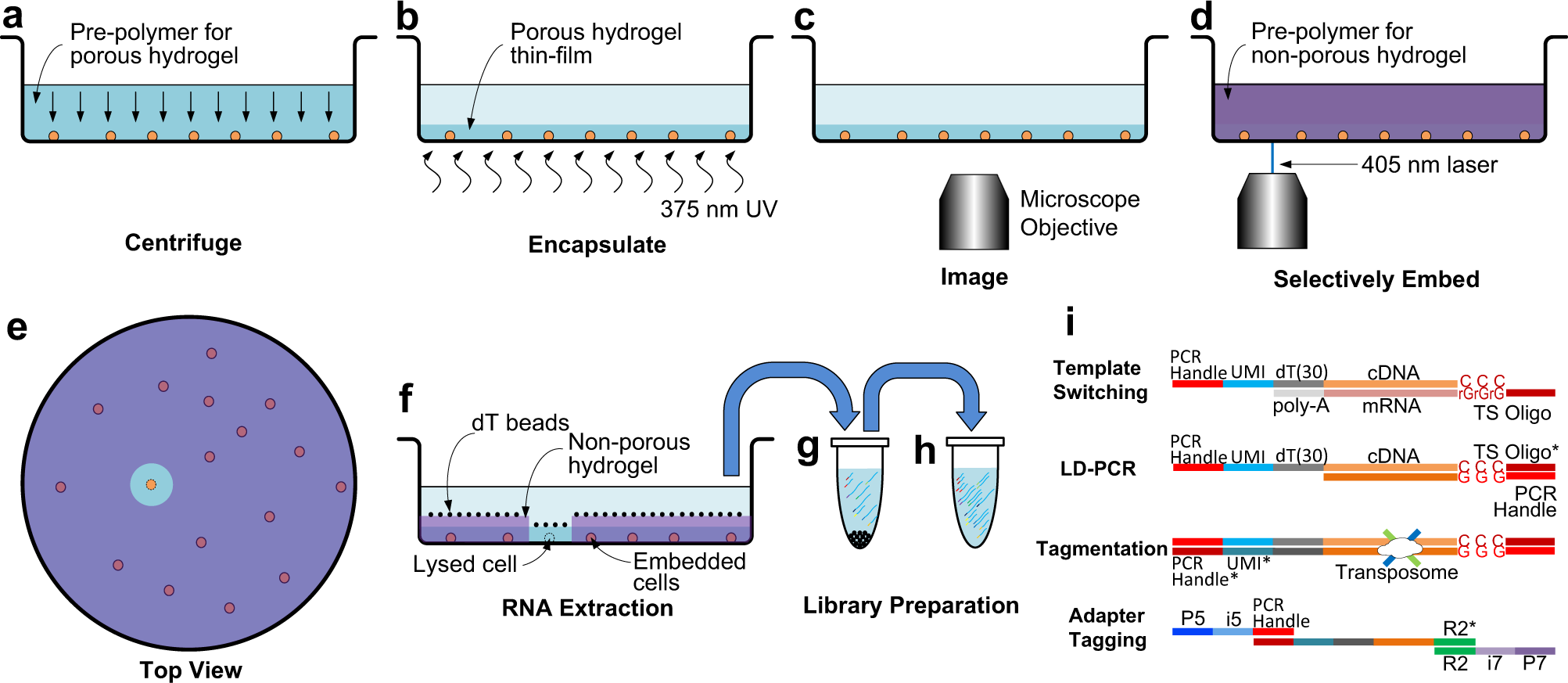
a The cell sample is distributed in imaging microwell plates with the pre-polymer for the porous hydrogel and aligned by centrifuging. b The cells are encapsulated in a porous hydrogel thin-film formed by exposure to 375 nm UV light. c Encapsulated cells are imaged by microscopy to identify a target cell in each well. d The pre-polymer for the non-porous hydrogel penetrates inside the porous hydrogel and is micropatterned using a scanning 405 nm laser. e (Top view) All cells except for the target cell are embedded in the non-porous hydrogel. f mRNA from the target cell is captured by oligo-dT beads. g The oligo-dT beads are transferred to a tube where mRNA is eluted, UMI-barcoded, and reverse transcribed with a template switching oligo. h The cDNA library is amplified by long distance PCR (LD-PCR). i Library preparation steps include template switching, LD-PCR, tagmentation, and adaptor tagging.
