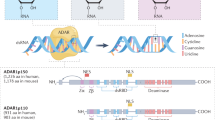
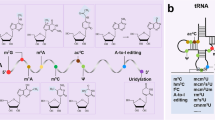
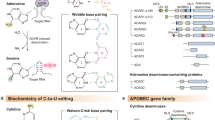
Abstract
Post-transcriptional RNA modifications can alter RNA structure, stability, localization, and function. Adenosine-to-inosine (A-to-I) RNA editing is a post-transcriptional modification that converts adenosine nucleotides in RNA to inosine nucleotides, catalyzed by adenosine-deaminase-acting-on-RNA (ADAR) enzymes. Recent studies have shown that A-to-I RNA editing is required for cardiovascular development and homeostasis whilst aberrant RNA editing plays a role in cardiovascular diseases. This article provides an overview of A-to-I RNA editing events that have been implicated in cardiovascular biology and disease. It also discusses harnessing RNA editing for cardiovascular disease biomarker development and engineering RNA editing for cardiovascular disease treatment.
Similar content being viewed by others
References
Benne, R. et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826 (1986).
Wagner, R. W., Smith, J. E., Cooperman, B. S. & Nishikura, K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. USA 86, 2647–2651 (1989).
Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016).
Heraud-Farlow, J. E. & Walkley, C. R. The role of RNA editing by ADAR1 in prevention of innate immune sensing of self-RNA. J. Mol. Med. 94, 1095–1102 (2016).
Jonkhout, N. et al. The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017).
Li, Q. et al. RNA editing underlies genetic risk of common inflammatory diseases. Nature 608, 569–577 (2022).
Blanc, V. & Davidson, N. O. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 594–602 (2010).
Teng, B., Burant, C. F. & Davidson, N. O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260, 1816–1819 (1993).
Blanc, V., Xie, Y., Luo, J., Kennedy, S. & Davidson, N. O. Intestine-specific expression of Apobec-1 rescues apolipoprotein B RNA editing and alters chylomicron production in Apobec1 -/- mice. J. Lipid Res. 53, 2643–2655 (2012).
Johannesen, C. D. L., Langsted, A., Nordestgaard, B. G. & Mortensen, M. B. Excess apolipoprotein B and cardiovascular risk in women and men. J. Am. Coll. Cardiol. 83, 2262–2273 (2024).
Sharma, S. et al. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 6, 6881 (2015).
Libby, P. Inflammation in atherosclerosis. Nature 420, 868–874 (2002).
Wang, I. X. et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 6, 906–915 (2014).
Kim, U., Wang, Y., Sanford, T., Zeng, Y. & Nishikura, K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. USA 91, 11457–11461 (1994).
Melcher, T. et al. A mammalian RNA editing enzyme. Nature 379, 460–464 (1996).
Chen, C. X. et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6, 755–767 (2000).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Zambrano-Mila, M. S. et al. Dissecting the basis for differential substrate specificity of ADAR1 and ADAR2. Nat. Commun. 14, 8212 (2023).
Tan, M. H. et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017).
Weldy, C. S. et al. Smooth muscle expression of RNA editing enzyme ADAR1 controls activation of the RNA sensor MDA5 in atherosclerosis. Nat. Cardiovasc. Res. 4, 1241–1257 (2025).
Wu, X. et al. ADAR2 increases in exercised heart and protects against myocardial infarction and doxorubicin-induced cardiotoxicity. Mol. Ther. 30, 400–414 (2022).
Gatsiou, A. et al. The RNA editor ADAR2 promotes immune cell trafficking by enhancing endothelial responses to interleukin-6 during sterile inflammation. Immunity 56, 979–997 e911 (2023).
Raghava Kurup, R. et al. RNA binding by ADAR3 inhibits adenosine-to-inosine editing and promotes expression of immune response protein MAVS. J. Biol. Chem. 298, 102267 (2022).
Oakes, E., Anderson, A., Cohen-Gadol, A. & Hundley, H. A. Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit B Pre-mRNA inhibits RNA editing in glioblastoma. J. Biol. Chem. 292, 4326–4335 (2017).
Patterson, J. B. & Samuel, C. E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388 (1995).
Heale, B. S. et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 28, 3145–3156 (2009).
Wang, Q. et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279, 4952–4961 (2004).
Liddicoat, B. J. et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015).
Moore, J. B. T. et al. The A-to-I RNA editing enzyme Adar1 is essential for normal embryonic cardiac growth and development. Circ. Res. 127, 550–552 (2020).
Garcia-Gonzalez, C. et al. ADAR1 prevents autoinflammatory processes in the heart mediated by IRF7. Circ. Res 131, 580–597 (2022).
Guo, X. et al. ADAR1 RNA editing regulates endothelial cell functions via the MDA-5 RNA sensing signaling pathway. Life Sci. Alliance 5, https://doi.org/10.26508/lsa.202101191 (2022).
Cai, D. & Chen, S. Y. ADAR1 is essential for smooth muscle homeostasis and vascular integrity. Cells 13, https://doi.org/10.3390/cells13151257 (2024).
Gerber, A., O’Connell, M. A. & Keller, W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA 3, 453–463 (1997).
Higuchi, M. et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000).
Mladenova, D. et al. Adar3 is involved in learning and memory in mice. Front. Neurosci. 12, 243 (2018).
Porath, H. T., Knisbacher, B. A., Eisenberg, E. & Levanon, E. Y. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 18, 185 (2017).
Bazak, L. et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 (2014).
Franzen, O. et al. Global analysis of A-to-I RNA editing reveals association with common disease variants. PeerJ 6, e4466 (2018).
Schaffer, A. A. et al. The cell line A-to-I RNA editing catalogue. Nucleic Acids Res. 48, 5849–5858 (2020).
Pinto, Y., Cohen, H. Y. & Levanon, E. Y. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15, R5 (2014).
Mann, T. D., Kopel, E., Eisenberg, E. & Levanon, E. Y. Increased A-to-I RNA editing in atherosclerosis and cardiomyopathies. PLoS Comput. Biol. 19, e1010923 (2023).
Lo Giudice, C., Tangaro, M. A., Pesole, G. & Picardi, E. Investigating RNA editing in deep transcriptome datasets with REDItools and REDIportal. Nat. Protoc. 15, 1098–1131 (2020).
Roth, S. H., Levanon, E. Y. & Eisenberg, E. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods 16, 1131–1138 (2019).
Kokot, K. E. et al. Reduction of A-to-I RNA editing in the failing human heart regulates formation of circular RNAs. Basic Res. Cardiol. 117, 32 (2022).
Shi, G. P. et al. Deficiency of the cysteine protease cathepsin S impairs microvessel growth. Circ. Res. 92, 493–500 (2003).
Sukhova, G. K. et al. Deficiency of cathepsin S reduces atherosclerosis in LDL receptor-deficient mice. J. Clin. Invest 111, 897–906 (2003).
Stellos, K. et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 22, 1140–1150 (2016).
Stossel, T. P. et al. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2, 138–145 (2001).
Levanon, E. Y. et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 33, 1162–1168 (2005).
Jain, M. et al. A-to-I RNA editing of Filamin A regulates cellular adhesion, migration and mechanical properties. FEBS J. 289, 4580–4601 (2022).
Jain, M. et al. RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure. EMBO J. 37, https://doi.org/10.15252/embj.201694813 (2018).
Kalayci, A. et al. Echocardiographic assessment of insulin-like growth factor binding protein-7 and early identification of acute heart failure. ESC Heart Fail. 7, 1664–1675 (2020).
Zhang, L. et al. Insulin-like growth factor-binding protein-7 (IGFBP7) links senescence to heart failure. Nat. Cardiovasc. Res. 1, 1195–1214 (2022).
Chen, R., McVey, D. G., Shen, D., Huang, X. & Ye, S. Phenotypic switching of vascular smooth muscle cells in atherosclerosis. J. Am. Heart Assoc. 12, e031121 (2023).
Fei, J., Cui, X. B., Wang, J. N., Dong, K. & Chen, S. Y. ADAR1-mediated RNA editing, a novel mechanism controlling phenotypic modulation of vascular smooth muscle cells. Circ. Res. 119, 463–469 (2016).
Vlachogiannis, N. I. et al. Adenosine-to-inosine Alu RNA editing controls the stability of the pro-inflammatory long noncoding RNA NEAT1 in atherosclerotic cardiovascular disease. J. Mol. Cell. Cardiol. 160, 111–120 (2021).
Boon, R. A. et al. MicroRNA-34a regulates cardiac ageing and function. Nature 495, 107–110 (2013).
Sygitowicz, G. & Sitkiewicz, D. Involvement of circRNAs in the development of heart failure. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms232214129 (2022).
Karagianni, K. et al. Recommendations for detection, validation, and evaluation of RNA editing events in cardiovascular and neurological/neurodegenerative diseases. Mol. Ther. Nucleic Acids 35, 102085 (2024).
Lee, J. H., Ang, J. K. & Xiao, X. Analysis and design of RNA sequencing experiments for identifying RNA editing and other single-nucleotide variants. RNA 19, 725–732 (2013).
Bahn, J. H. et al. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 22, 142–150 (2012).
Ramaswami, G. et al. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 10, 128–132 (2013).
Srivastava, P. K. et al. Genome-wide analysis of differential RNA editing in epilepsy. Genome Res. 27, 440–450 (2017).
Diroma, M. A., Ciaccia, L., Pesole, G. & Picardi, E. Elucidating the editome: bioinformatics approaches for RNA editing detection. Brief. Bioinform. 20, 436–447 (2019).
Uchida, S. & Jones, S. P. RNA editing: unexplored opportunities in the cardiovascular system. Circ. Res 122, 399–401 (2018).
D’Addabbo, P. et al. REDIportal: toward an integrated view of the A-to-I editing. Nucleic Acids Res. 53, D233–D242 (2025).
Piechotta, M., Naarmann-de Vries, I. S., Wang, Q., Altmuller, J. & Dieterich, C. RNA modification mapping with JACUSA2. Genome Biol. 23, 115 (2022).
Tran, S. S., Zhou, Q. & Xiao, X. Statistical inference of differential RNA-editing sites from RNA-sequencing data by hierarchical modeling. Bioinformatics 36, 2796–2804 (2020).
Torkler, P. et al. LoDEI: a robust and sensitive tool to detect transcriptome-wide differential A-to-I editing in RNA-seq data. Nat. Commun. 15, 9121 (2024).
Lerner, T. et al. C-to-U RNA editing: from computational detection to experimental validation. Methods Mol. Biol. 2181, 51–67 (2021).
van der Kwast, R. et al. Adenosine-to-inosine editing of microRNA-487b alters target gene selection after ischemia and promotes neovascularization. Circ. Res. 122, 444–456 (2018).
Wang, F. et al. Lessons from discovery of true ADAR RNA editing sites in a human cell line. BMC Biol. 21, 160 (2023).
van der Kwast, R. et al. Adenosine-to-inosine editing of vasoactive microRNAs alters their targetome and function in ischemia. Mol. Ther. Nucleic Acids 21, 932–953 (2020).
Voss, G. & Ceder, Y. Two-tailed RT-qPCR for the auantification of A-to-I-edited microRNA isoforms. Curr. Protoc. 3, e645 (2023).
Pfeiffer, L. S. & Stafforst, T. Precision RNA base editing with engineered and endogenous effectors. Nat. Biotechnol. 41, 1526–1542 (2023).
Zhang, D., Zhu, L., Gao, Y., Wang, Y. & Li, P. RNA editing enzymes: structure, biological functions and applications. Cell Biosci. 14, 34 (2024).
Qu, L. et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 37, 1059–1069 (2019).
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
Birgaoanu, M., Sachse, M. & Gatsiou, A. RNA editing therapeutics: advances, challenges and perspectives on combating heart disease. Cardiovasc. Drugs Ther. 37, 401–411 (2023).
Porto, E. M., Komor, A. C., Slaymaker, I. M. & Yeo, G. W. Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 19, 839–859 (2020).
Musunuru, K. Moving toward genome-editing therapies for cardiovascular diseases. J. Clin. Investig. 132, https://doi.org/10.1172/JCI148555 (2022).
Musunuru, K. et al. Patient-specific in vivo gene editing to treat a rare genetic disease. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2504747 (2025).
Shinwari, Z. K., Tanveer, F. & Khalil, A. T. Ethical issues regarding CRISPR mediated genome editing. Curr. Issues Mol. Biol. 26, 103–110 (2018).
Yang, P. et al. Allele-specific suppression of variant MHC with high-precision RNA nuclease CRISPR-Cas13d prevents hypertrophic cardiomyopathy. Circulation 150, 283–298 (2024).
Mehta, A. & Shapiro, M. D. Apolipoproteins in vascular biology and atherosclerotic disease. Nat. Rev. Cardiol. 19, 168–179 (2022).
Mullard, A. RNA-editing drugs advance into clinical trials. Nat. Rev. Drug Discov. 23, 323–326 (2024).
De Chiara, F. et al. ADAR RNA editing for cardiovascular disease: targeting B4GALT1 to modulate lipid metabolism through reduced galatosyltransferase activity. bioRxiv. https://doi.org/10.1101/2025.10.30.685343.
Acknowledgements
S.Y. thanks funding from the British Heart Foundation (RG/16/13/32609, RG/19/9/34655, PG/16/9/31995, PG/18/73/34059, and SP/19/2/344612), the National Medical Research Council of Singapore (MOH-001229 and MOH-001479), and the National University of Singapore/National University Health System (NUHSRO/2022/004/Startup/01). C.U.S. was a Leicester British Heart Foundation Accelerator Award (AA/18/3/34220) Research Fellow. D.G.M. is supported by the British Heart Foundation Research Excellence Award (RE/24/130031), the van Geest Foundation Heart and Cardiovascular Diseases Research Fund, and was awarded a BHF Accelerator Early Careers Researcher Interdisciplinary Fellowship and pump-priming funding from the Leicester British Heart Foundation Accelerator Award (AA/18/3/34220). This work falls under the portfolio of research conducted within the National Institute for Health Research Leicester Biomedical Research Centre and the Leicester BHF Centre of Research Excellence.
Author information
Authors and Affiliations
Contributions
X.H. and S.Y. drafted the manuscript. C.U.S., D.G.M. and S.Y. revised the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Mengtan Xing and Christina Karlsson Rosenthal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, X., Solomon, C., McVey, D.G. et al. RNA editing in cardiovascular health and disease. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09680-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09680-1