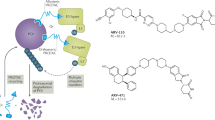
Abstract
PROteolysis TArgeting Chimeras (PROTACs) are bifunctional molecules that offer a novel approach to targeted protein degradation, showing particular promise for previously ‘undruggable’ targets. Despite their therapeutic potential, significant challenges remain in optimizing the pharmacokinetic (PK) properties of PROTACs, particularly in terms of their ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) characteristics. In this study, we drew on the experience of previous work and combined traditional machine learning with multiple molecular fingerprints to propose a PROTAC pharmacokinetic property prediction model EGFR-PROPK. We conducted in-vivo experiments on 100 EGFR-targeting PROTAC molecules, focusing on clearance (CL), half-life (T1/2), and apparent volume of distribution (Vss), which are the most intuitive and commonly used macro parameters to describe the PK properties of a drug since they describe the absorption-distribution-elimination process of drugs in the body from different perspectives. Our findings reveal that traditional models trained on small molecules perform poorly on PROTACs. However, training the models on PROTAC-specific data significantly improved prediction accuracy, achieving correlation coefficients of 0.78, 0.75, and 0.52 between the predicted and observed values for T1/2, CL and Vss, respectively, highlighting the need for tailored approaches in PK evaluation for these unique molecules. These insights are critical for advancing the design and development of PROTAC-based therapies.

Similar content being viewed by others
Data availability
The ADMET dataset ultilized for pre-training is sourced from the public accessible Therapeutics Data Commons (TDC), avaliable at https://tdcommons.ai/.
Code availability
All the data sets and source code are publicly available through the GitHub(https://github.com/Zhang-Ran-0119/EGFR-PROPK). The custom code used in this study is available at GitHub and has been archived on Zenodo with a persistent https://doi.org/10.5281/zenodo.18276159. The repository includes all scripts required to reproduce the reported results.
References
Bondeson, D. P. & Crews, C. M. Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 57, 107–123 (2017).
Schneider, M. et al. The protactable genome. Nat. Rev. Drug Discov. 20, 789–797 (2021).
Lai, A. C. & Crews, C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017).
Hammoudeh, D. I., Follis, A. V., Prochownik, E. V. & Metallo, S. J. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 131, 7390–7401 (2009).
An, S. & Fu, L. Small-molecule PROTACs: an emerging and promising approach for the development of targeted therapy drugs. EBioMedicine 36, 553–562 (2018).
Burslem, G. M. & Crews, C. M. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181, 102–114 (2020).
Nowak, R. P. et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 14, 706–714 (2018).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 98, 8554–8559 (2001).
Weng, G. et al. PROTAC-DB: an online database of PROTACs. Nucleic Acids Res. 49, D1381–D1387 (2021).
Weng, G. et al. PROTAC-DB 2.0: an updated database of PROTACs. Nucleic Acids Res. 51, D1367–D1372 (2023).
Ge, J. et al. PROTAC-DB 3.0: an updated database of PROTACs with extended pharmacokinetic parameters. Nucleic Acids Res. 53, 1510–1515 (2024).
Bai F., Tian S., Tang Y., Li F. & Li Z. PROTAC-Databank: the present largest integrated resource of PROTACs, enabling the enhanced DeepPROTACs 2.0 for degradation prediction. (2024).
Zheng, S. et al. Accelerated rational PROTAC design via deep learning and molecular simulations. Nat. Mach. Intell. 4, 739–748 (2022).
Li B., Ran T. & Chen H. 3D based generative PROTAC linker design with reinforcement learning. Brief. Bioinf. 24, bbad323 (2023).
Li, F. et al. DeepPROTACs is a deep learning-based targeted degradation predictor for PROTACs. Nat. Commun. 13, 7133 (2022).
Li F., Hu Q., Zhou Y., Yang H. & Bai F. DiffPROTACs is a deep learning-based generator for proteolysis targeting chimeras. Brief. Bioinf. 25, bbae358 (2024).
Wang, X. et al. Annual review of PROTAC degraders as anticancer agents in 2022. Eur. J. Med. Chem. 267, 116166 (2024).
Swanson, K. et al. ADMET-AI: a machine learning ADMET platform for evaluation of large-scale chemical libraries. Bioinformatics 40, 573531 (2024).
Wei, Y., Li, S., Li, Z., Wan, Z. & Lin, J. Interpretable-ADMET: a web service for ADMET prediction and optimization based on deep neural representation. Bioinformatics 38, 2863–2871 (2022).
Fu, L. et al. ADMETlab 3.0: an updated comprehensive online ADMET prediction platform enhanced with broader coverage, improved performance, API functionality and decision support. Nucleic Acids Res. 52, W422–W431 (2024).
Huang, K. et al. Therapeutics data commons: machine learning datasets and tasks for drug discovery and development. Proc. Neural Inf. Process. Syst. Track Datasets Benchmarks 1, 09548 (2021).
Notwell J. H. & Wood M. W. ADMET property prediction through combinations of molecular fingerprints. arXiv (2023).
Tian, H., Ketkar, R. & Tao, P. ADMETboost: a web server for accurate ADMET prediction. J. Mol. Model 28, 408 (2022).
Yang, H. et al. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 35, 1067–1069 (2018).
Peteani, G., Huynh, M. T. D., Gerebtzoff, G. & Rodríguez-Pérez, R. Application of machine learning models for property prediction to targeted protein degraders. Nat. Commun. 15, 5764 (2024).
Passaro, A., Jänne, P. A., Mok, T. & Peters, S. Overcoming therapy resistance in EGFR-mutant lung cancer. Nat. Cancer 2, 377–391 (2021).
Growth factor receptor expression in anal squamous lesions: modifications associated with oncogenic human papillomavirus and human immunodeficiency virus. Human Pathol. 40, 1517–1527 (2009).
An, Z., Aksoy, O., Zheng, T., Fan, Q.-W. & Weiss, W. A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene 37, 1561–1575 (2018).
Alharbi, K. S. et al. An overview of epithelial growth factor receptor (EGFR) inhibitors in cancer therapy. Chem. Biol. Interact. 366, 110108 (2022).
Zhen, Y., Guanghui, L. & Xiefu, Z. Knockdown of EGFR inhibits growth and invasion of gastric cancer cells. Cancer Gene Ther. 21, 491–497 (2014).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Gridelli, C. et al. Erlotinib in non-small cell lung cancer treatment: current status and future development. Oncologist 12, 840–849 (2007).
Jiang, Y. et al. Afatinib for the treatment of NSCLC with uncommon EGFR mutations: a narrative review. Curr. Oncol. 30, 5337–5349 (2023).
Liang, W. et al. Network meta-analysis of erlotinib, gefitinib, afatinib and icotinib in patients with advanced non-small-cell lung cancer harboring EGFR mutations. PLoS ONE 9, e85245 (2014).
Li, Q.-H. et al. Anti-EGFR therapy in metastatic colorectal cancer: mechanisms and potential regimens of drug resistance. Gastroenterol. Rep. 8, 179–191 (2020).
Du, Y. et al. HJM-561, a potent, selective, and orally bioavailable EGFR PROTAC that overcomes osimertinib-resistant EGFR triple mutations. Mol. Cancer Ther. 21, 1060–1066 (2022).
ZHANG, Y. G. et al. Class of novel protein degradation agents and application thereof. (WIPO, 2022).
YE, G. et al. Protein inhibitor or degrading agent, pharmaceutical composition containing same and pharmaceutical use. (WIPO, 2022).
Prokhorenkova L., Gusev G., Vorobev A., Dorogush A. V. & Gulin A. CatBoost: unbiased boosting with categorical features. In: Proceedings of the 32nd International Conference on Neural Information Processing Systems. (Curran Associates Inc., 2018).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Gedeck, P., Rohde, B. & Bartels, C. QSAR − How good is it in practice? Comparison of descriptor sets on an unbiased cross section of corporate data sets. J. Chem. Inf. Model. 46, 1924–1936 (2006).
Stiefl, N., Watson, I. A., Baumann, K. & Zaliani, A. ErG: 2D pharmacophore descriptions for scaffold hopping. J. Chem. Inf. Model. 46, 208–220 (2006).
Hu, W. et al. Strategies for pre-training graph neuralnetworks. arXiv:190512265, (2020).
Talevi A. & Bellera C. L. Total Clearance and Organ Clearance. In: The ADME Encyclopedia: a Comprehensive Guide on Biopharmacy and Pharmacokinetics. (Springer International Publishing, 2021).
Lombardo, F., Obach, R. S., Shalaeva, M. Y. & Gao, F. Prediction of volume of distribution values in humans for neutral and basicdrugs using physicochemical measurements and plasma protein binding data. J. Med. Chem. 45, 10 (2002).
Grime, K. & Riley, R. J. The impact of in vitro binding on in vitro-in vivo extrapolations, projections of metabolic clearance and clinical drug-drug interactions. Curr. Drug Metab. 7, 251–264 (2006).
Obach, R. S. et al. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J. Pharmacol. Exp. Therapeut. 283, 46–58 (1997).
Benet, L. Z. & Zia-Amirhosseini, P. Basic principles of pharmacokinetics. Toxicol. Pathol. 23, 115–123 (1995).
Acknowledgements
This work was supported by National Key R&D Program of China (to F. B., Grant IDs: 2022YFC3400501 and 2022YFC3400500), Shanghai Science and Technology Development Fund (to F. B., 22ZR1441400), Shanghai Pujiang Program (to Y. L., Grant No. 23PJ1420700), ShanghaiTech AI4S Initiative SHTAI4S202404, start-up package from ShanghaiTech University, and Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine at ShanghaiTech University. We are grateful for the support from HPC Platform of ShanghaiTech University.
Author information
Authors and Affiliations
Contributions
R.Z. and F.L. constructed and trained the network model and wrote the manuscript; Q.Z., Y.L., Y.G., and L.Z. conducted the experiments and provide data; F.B. and L.Z. designed the whole project and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose. Fang Bai is an Editorial Board Member for Communications Chemistry, but was not involved in the editorial review of, or the decision to publish this article.
Peer review
Peer review information
Communications Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, R., Li, F., Liu, Y. et al. A machine learning-based pharmacokinetics predictor (EGFR-PROPK) for EGFR-targeting PROTACs. Commun Chem (2026). https://doi.org/10.1038/s42004-026-01938-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-026-01938-3