Abstract
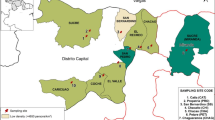
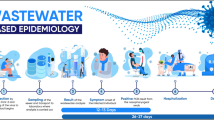
Wastewater surveillance can track trends in multiple pathogens simultaneously by leveraging efficient laboratory processing. In Switzerland, wastewater surveillance of four respiratory pathogens is conducted at 14 locations representing 2.3 million people. Trends in respiratory diseases are tracked using a six-plex digital PCR assay targeting influenza A, influenza B, respiratory syncytial virus and SARS-CoV-2 N1 and N2 regions and murine hepatitis virus for recovery efficiency control. Wastewater data were integrated with disease data from two reporting systems, and comparisons from July 2023 to July 2024 showed strong agreement for most targets. Lower correspondence for influenza B highlighted challenges in tracking disease dynamics during seasons without pronounced outbreaks. Wastewater monitoring further revealed that targeting N1 or N2 led to divergent estimates of SARS-CoV-2 loads, highlighting the impact of mutations in assay target regions. The study emphasizes the importance of an integrated wastewater monitoring programme as a complementary tool for public health surveillance, demonstrating clear concordance with clinical data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Digital PCR data are available for download from wise.ethz.ch. Only data that passed our described quality control processes are available here. All data are available on request. Weekly case data are available from the Federal Office of Public Health (FOPH) via a public API (https://api.idd.bag.admin.ch/swagger-ui/api-doc.html). Daily case data were specifically requested from the FOPH.
Code availability
All code used in the analysis is available via GitHub at https://github.com/EawagPHH/A-six-plex-digital-PCR-assay-for-monitoring-respiratory-viruses-in-wastewater/blob/main/Analysis.Rmd.
References
Troeger, C. et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 17, 1133–1161 (2017).
Hirve, S. et al. Influenza seasonality in the tropics and subtropics - when to vaccinate? PLoS ONE 11, e0153003 (2016).
Reed, C. et al. Estimating influenza disease burden from population-based surveillance data in the United States. PLoS ONE 10, e0118369 (2015).
Xagoraraki, I. & O’Brien, E. in Women in Water Quality: Investigations by Prominent Female Engineers (ed. Deborah Jean O’Bannon) 75–97 (Springer International Publishing, 2020).
Sims, N. & Kasprzyk-Hordern, B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 139, 105689 (2020).
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R. & Brouwer, A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 7, 511–516 (2020).
Medema, G., Been, F., Heijnen, L. & Petterson, S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health 17, 49–71 (2020).
Fernandez-Cassi, X. et al. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. Water Res. 200, 117252 (2021).
Peccia, J. et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 38, 1164–1167 (2020).
Kilaru, P. et al. Wastewater surveillance for infectious disease: a systematic review. Am. J. Epidemiol. 192, 305–322 (2022).
Hughes, B. et al. Respiratory syncytial virus (RSV) RNA in wastewater settled solids reflects RSV clinical positivity rates. Environ. Sci. Technol. Lett. 9, 173–178 (2022).
Wolfe, M. K. et al. Wastewater-based detection of two influenza outbreaks. Environ. Sci. Technol. Lett. 9, 687–692 (2022).
Zheng, X. et al. Development and application of influenza virus wastewater surveillance in Hong Kong. Water Res. 245, 120594 (2023).
Whale, A. S., Huggett, J. F. & Tzonev, S. Fundamentals of multiplexing with digital PCR. Biomol. Detect. Quantif. 10, 15–23 (2016).
Nadeau, S. et al. Influenza transmission dynamics quantified from RNA in wastewater in Switzerland. Swiss Med. Wkly 154, 3503 (2024).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. 4th edn (Springer New York, 2002).
Huang, Z. et al. Development and validation of an automated and high-throughput quadruplex RT-ddPCR assay for the detection of influenza A, influenza B, respiratory syncytial virus, and SARS-CoV-2. Front. Cell. Infect. Microbiol. 15, 1529336 (2025).
Zafeiriadou, A., Kaltsis, L., Thomaidis, N. S. & Markou, A. Simultaneous detection of influenza A, B and respiratory syncytial virus in wastewater samples by one-step multiplex RT-ddPCR assay. Hum. Genom. 18, 48 (2024).
WISE Dashboard–Wastewater-based Infectious Disease Surveillance and Epidemiology. ETH Zurich and Eawag https://wise.ethz.ch/ (2025).
Zheng, X. et al. Comparison of virus concentration methods and RNA extraction methods for SARS-CoV-2 wastewater surveillance. Sci. Total Environ. 824, 153687 (2022).
Ahmed, W. et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 739, 139960 (2020).
Hart, J. J., Jamison, M. N., McNair, J. N. & Szlag, D. C. Frequency and degradation of SARS-CoV-2 markers N1, N2, and E in sewage. J. Water Health 21, 514–524 (2023).
Sun, J. et al. Underestimation of SARS-CoV-2 in wastewater due to single or double mutations in the N1 qPCR probe binding region. Water Res. X 22, 100221 (2024).
Thakali, O. et al. Real-time evaluation of signal accuracy in wastewater surveillance of pathogens with high rates of mutation. Sci. Rep. 14, 3728 (2024).
Chen, C. et al. CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics 38, 1735–1737 (2021).
Huisman, J. S. et al. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. Environ. Health Perspect. 130, 057011 (2022).
Similarities and differences between flu and COVID-19. CDC https://www.cdc.gov/flu/about/flu-vs-covid19.html (2024).
Aliza Rosen, M. H. What to know about JN.1, the latest Omicron variant. Johns Hopkins Bloomberg School of Public Health https://publichealth.jhu.edu/2024/jn1-the-dominant-variant-in-the-covid-surge (2024).
Idris, I. & Adesola, R. O. Emergence and spread of JN.1 COVID-19 variant. Bull. Natl Res. Cent. 48, 27 (2024).
Kaku, Y. et al. Virological characteristics of the SARS-CoV-2 KP.2 variant. bioRxiv https://doi.org/10.1101/2024.04.24.590786 (2024).
Faherty, E. A. G. et al. Correlation of wastewater surveillance data with traditional influenza surveillance measures in Cook County, Illinois, October 2022–April 2023. Sci. Total Environ. 912, 169551 (2024).
Wolken, M. et al. Wastewater surveillance of SARS-CoV-2 and influenza in preK-12 schools shows school, community, and citywide infections. Water Res. 231, 119648 (2023).
Wade, M. J. et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 424, 127456 (2022).
Ahmed, W. et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 761, 144216 (2021).
Wu, F. et al. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 805, 150121 (2022).
Weidhaas, J. et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 775, 145790 (2021).
Rezaeitavabe, F. et al. Beyond linear regression: modeling COVID-19 clinical cases with wastewater surveillance of SARS-CoV-2 for the city of Athens and Ohio University campus. Sci. Total Environ. 912, 169028 (2024).
Group, T. C. & Huggett, J. F. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 66, 1012–1029 (2020).
Stokdyk, J. P., Firnstahl, A. D., Spencer, S. K., Burch, T. R. & Borchardt, M. A. Determining the 95% limit of detection for waterborne pathogen analyses from primary concentration to qPCR. Water Res. 96, 105–113 (2016).
Leibowitz, J., Kaufman, G. & Liu, P. Coronaviruses: propagation, quantification, storage, and construction of recombinant mouse hepatitis virus. Curr. Protoc. Microbiol. 21, 1511–15146 (2011).
Mandatory reporting system. FOPH https://idd.bag.admin.ch/survey-systems/oblig (2025).
Acknowledgements
We would like to thank present and past members of the Laboratory Monitoring Team for developing the methods, processing samples and generating and analysing data. Additionally, we thank present and past collaborators of the WISE (Wastewater-based Infectious disease Surveillance and Epidemiology) project for their support and valuable intellectual contributions and NEXUS for their setting up and supporting the WISE database (wisedb.ethz.ch) which facilitated our analysis. We thank the technical staff of each WWTP for promptly sending us the wastewater samples on a weekly basis, the FOPH for sharing case data with us and A.C. Ripolles and T. Kohn for providing us with MHV. This study was funded by the Swiss National Science Foundation (SNSF Sinergia grant number CRSII5_205933) and by the Swiss Federal Office of Public Health (grant numbers 142006108/334.0-101/26 and 142006655/334.0-107/12) granted to C.O. and T.R.J.
Author information
Authors and Affiliations
Contributions
C.O. and T.R.J. contributed equally. C.O. and T.R.J. conceptualized, supervised and provided funding. M.P. and R.E.M. designed the study, performed analyses, prepared visualizations and wrote the first draft of the manuscript. L.C., J.d.K.-E., C.G. and S.K. contributed invaluable scientific insights. P.S. provided statistical support. A.D., C.H., A.H., G.L., L.S., A.W. and D.Y. helped with sample processing, data generation and curation. We declare that all authors have seen and approved the manuscript and have contributed substantially to the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Water thanks Robert Delatolla, David Walker and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–6, Figs. 1–5 and Texts 1 and 2.
Supplementary Table 1
dMIQE2020 checklist for authors, reviewers and editors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pitton, M., McLeod, R.E., Caduff, L. et al. A six-plex digital PCR assay for monitoring respiratory viruses in wastewater. Nat Water 3, 1174–1186 (2025). https://doi.org/10.1038/s44221-025-00503-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44221-025-00503-x