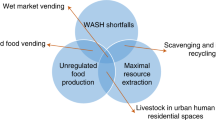
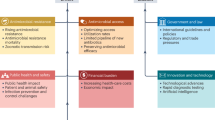
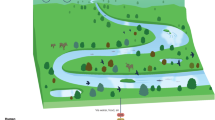
Abstract
The first therapeutic use of antimicrobial agents initiated their endless arms race with antimicrobial resistance (AMR). Although the genes encoding antimicrobial resistance are ancient and ubiquitous in various environmental compartments, including aquatic environments, over eight decades of exposure to selective pressure has changed the way antimicrobial resistance genes (ARGs) emerge and transmit among the three One Health sectors (that is, the intersected sectors of humans, animals and the environment). The dissemination of ARGs has been facilitated by the widespread use of antimicrobials, along with direct and secondary pollution pathways. Current global consensus dictates that AMR should be addressed under a One Health framework. AMR National Action Plans have frequently been formulated. However, the capacity for implementation is not ready in most countries, especially in low- and middle-income regions. This is in part due to the substantial challenges in documenting and controlling cross-sector AMR connectivity. Here we describe the past and current status of AMR, emphasizing the contribution of connectivity to global AMR burden. We discuss connectivity at ecological, microbial and genetic levels; propose an approach based on genomics and metagenomics to assess connectivity; and finally advocate for cross-sector studies to better understand AMR connectivity and mitigate dissemination. We believe that such harmonized connectivity studies will facilitate coordinated actions and investments across sectors and regions to scale up AMR management globally.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Towards Specific Commitments and Action in the Response to Antimicrobial Resistance (Global Leaders Group (GLG) on Antimicrobial Resistance, 2024); https://www.amrleaders.org/about-us/what-we-do/glg-report
Larsson, D. G. J. & Flach, C. F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269 (2022).
Berendonk, T. U. et al. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317 (2015).
One Health Initiative (World Health Organization); https://www.who.int/teams/one-health-initiative/quadripartite-secretariat-for-one-health
Velazquez-Meza, M. E., Galarde-López, M., Carrillo-Quiróz, B. & Alpuche-Aranda, C. M. Antimicrobial resistance: One Health approach. Vet. World 15, 743–749 (2022).
McEwen, S. A. & Collignon, P. J. Antimicrobial resistance: a One Health perspective. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.arba-0009-2017 (2018).
Arnold, K. E. et al. The need for One Health systems-thinking approaches to understand multiscale dissemination of antimicrobial resistance. Lancet Planet. Health 8, e124–e133 (2024).
Hernando-Amado, S., Coquet, T. M., Baquero, F. & Martinez, J. L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442 (2019).
Jin, L., Xie, J., He, T., Wu, D. & Li, X. Airborne transmission as an integral environmental dimension of antimicrobial resistance through the ‘One Health’ lens. Crit. Rev. Environ. Sci. Technol. 52, 4172–4193 (2022).
Amarasiri, M., Sano, D. & Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 50, 2016–2059 (2020).
Manaia, C. M. et al. Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ. Int. 115, 312–324 (2018).
Chen, C. et al. Characterising global antimicrobial resistance research explains why One Health solutions are slow in development: an application of AI-based gap analysis. Environ. Int. 187, 108680 (2024).
Pehrsson, E. C. et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216 (2016).
Maciel-Guerra, A. et al. Dissecting microbial communities and resistomes for interconnected humans, soil and livestock. ISME J. 17, 21–35 (2023).
Marshall, D. A. et al. Impact of antibiotic administrative restrictions on trends in antibiotic resistance. Can. J. Public Health 97, 126–131 (2006).
Davies, J. & Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 (2010).
Kirchhelle, C. Swann song: antibiotic regulation in British livestock production (1953–2006). Bull. Hist. Med. 92, 317–350 (2018).
Barton, M. D. Antibiotic use in animal feed and its impact on human health. Nutr. Res. Rev. 13, 279–299 (2000).
Andersson, D. I. & Hughes, D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271 (2010).
Durao, P., Balbontin, R. & Gordo, I. Evolutionary mechanisms shaping the maintenance of antibiotic resistance. Trends Microbiol. 26, 677–691 (2018).
Allel, K. et al. Global antimicrobial-resistance drivers: an ecological country-level study at the human-animal interface. Lancet Planet. Health 7, e291–e303 (2023).
Kim, D. W. & Cha, C. J. Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 53, 301–309 (2021).
Holmes, A. H. et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187 (2016).
Liu, Y. Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168 (2016).
Wang, R. B. et al. The global distribution and spread of the mobilized colistin resistance gene. Nat. Commun. 9, 1179 (2018).
González, S. O., Almeida, C. A., Calderón, M., Mallea, M. A. & González, P. Assessment of the water self-purification capacity on a river affected by organic pollution: application of chemometrics in spatial and temporal variations. Environ. Sci. Pollut. Res. Int. 21, 10583–10593 (2014).
Xie, Y. et al. Insight into impact of sewage discharge on microbial dynamics and pathogenicity in river ecosystem. Sci. Rep. 12, 6894 (2022).
Knapp, C. W., Dolfing, J., Ehlert, P. A. I. & Graham, D. W. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44, 580–587 (2010).
Zhao, Y. et al. Global soil antibiotic resistance genes are associated with increasing risk and connectivity to human resistome. Nat. Commun. 16, 7141 (2025).
Wilkinson, J. L. et al. Pharmaceutical pollution of the world’s rivers. Proc. Natl Acad. Sci. USA 119, e2113947119 (2022).
Tozer, L. Water pollution ‘timebomb’ threatens global health. Nature https://doi.org/10.1038/d41586-023-02337-7 (2023).
Wang, X. W. et al. Ecological dynamics imposes fundamental challenges in community-based microbial source tracking. iMeta 2, e75 (2023).
Li, L. G., Huang, Q., Yin, X. L. & Zhang, T. Source tracking of antibiotic resistance genes in the environment—challenges, progress and prospects. Water Res. 185, 116127 (2020).
MacLean, R. C. & San Millan, A. The evolution of antibiotic resistance. Science 365, 1082–1083 (2019).
Pal, A. & Andersson, D. I. Bacteria can compensate the fitness costs of amplified resistance genes via a bypass mechanism. Nat. Commun. 15, 2333 (2024).
Che, Y. et al. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl Acad. Sci. USA 118, e2008731118 (2021).
Djordjevic, S. P., Stokes, H. W. & Chowdhury, P. R. Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front. Microbiol. 4, 86 (2013).
Partridge, S. R., Kwong, S. M., Firth, N. & Jensen, S. O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088-17 (2018).
Gillings, M. R. et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279 (2015).
Botelho, J. & Schulenburg, H. The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 29, 8–18 (2021).
Wright, G. D. Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594 (2010).
Benveniste, R. & Davies, J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 70, 2276–2280 (1973).
Poirel, L., Kampfer, P. & Nordmann, P. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 46, 4038–4040 (2002).
Marshall, C. G., Lessard, I. A., Park, I. & Wright, G. D. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42, 2215–2220 (1998).
Pärnänen, K. M. M. et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5, eaau9124 (2019).
Lee, K. et al. Mobile resistome of human gut and pathogen drives anthropogenic bloom of antibiotic resistance. Microbiome 8, 2 (2020).
Nadimpalli, M. et al. Combating global antibiotic resistance: emerging One Health concerns in lower- and middle-income countries. Clin. Infect. Dis. 66, 963–969 (2018).
Baker-Austin, C., Wright, M. S., Stepanauskas, R. & McArthur, J. V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182 (2006).
Wales, A. D. & Davies, R. H. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604 (2015).
Manaia, C. M. et al. The complex interplay between antibiotic resistance and pharmaceutical and personal care products in the environment. Environ. Toxicol. Chem. 43, 637–652 (2024).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Wang, Y. et al. Antidepressants can induce mutation and enhance persistence toward multiple antibiotics. Proc. Natl Acad. Sci. USA 120, e2208344120 (2023).
Rodríguez-Molina, D. et al. International travel as a risk factor for carriage of extended-spectrum β-lactamase-producing in a large sample of European individuals - the AWARE study. Int. J. Env. Res. Public Health 19, 4758 (2022).
UNEP. Bracing for Superbugs: Strengthening Environmental Action in the One Health Response to Antimicrobial Resistance (United Nations Environment Programme, 2023); https://www.unep.org/resources/superbugs/environmental-action
Rodrigues, J. L. M. et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl Acad. Sci. USA 110, 988–993 (2013).
Gossner, M. M. et al. Land-use intensification causes multitrophic homogenization of grassland communities. Nature 540, 266–269 (2016).
Peixoto, R. S. et al. Harnessing the microbiome to prevent global biodiversity loss. Nat. Microbiol. 7, 1726–1735 (2022).
WHO. Global Action Plan on Antimicrobial Resistance (World Health Organization, 2015); https://www.who.int/publications/i/item/9789241509763
WHO. Library of AMR National Action Plans (World Health Organization, 2023); https://www.who.int/teams/surveillance-prevention-control-AMR/national-action-plan-monitoring-evaluation/library-of-national-action-plans
Ikhimiukor, O. O., Odih, E. E., Donado-Godoy, P. & Okeke, I. N. A bottom-up view of antimicrobial resistance transmission in developing countries. Nat. Microbiol. 7, 757–765 (2022).
WHO. Global Database for Tracking Antimicrobial Resistance (AMR) Country Self-Assessment Survey (TrACSS) (World Health Organization, 2023); https://amrcountryprogress.org/#/map-view
Gillings, M. R. Lateral gene transfer, bacterial genome evolution and the anthropocene. Ann. N. Y. Acad. Sci. 1389, 20–36 (2017).
Zhao, Y. et al. Antibiotic resistome in the livestock and aquaculture industries: status and solutions. Crit. Rev. Environ. Sci. Technol. 51, 2159–2196 (2021).
Rousham, E. K. et al. Human colonization with extended-spectrum beta-lactamase-producing E. coli in relation to animal and environmental exposures in Bangladesh: an observational One Health study. Environ. Health Perspect. 129, 37001 (2021).
Li, L. et al. Extended-spectrum beta-lactamase and carbapenemase genes are substantially and sequentially reduced during conveyance and treatment of urban sewage. Environ. Sci. Technol. 55, 5939–5949 (2021).
Worby, C. J. et al. Gut microbiome perturbation, antibiotic resistance and Escherichia coli strain dynamics associated with international travel: a metagenomic analysis. Lancet Microbe 4, e790–e799 (2023).
Gillings, M. R. & Paulsen, I. T. Microbiology of the Anthropocene. Anthropocene 5, 1–8 (2014).
Jordt, H. et al. Coevolution of host-plasmid pairs facilitates the emergence of novel multidrug resistance. Nat. Ecol. Evol. 4, 863–869 (2020).
Loftie-Eaton, W. et al. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 1, 1354–1363 (2017).
Portik, D. M., Brown, C. T. & Pierce-Ward, N. T. Evaluation of taxonomic classification and profiling methods for long-read shotgun metagenomic sequencing datasets. BMC Bioinformatics 23, 541 (2022).
Zolfo, M., Tett, A., Jousson, O., Donati, C. & Segata, N. MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res. 45, e7 (2017).
Zheng, W. et al. High-throughput, single-microbe genomics with strain resolution, applied to a human gut microbiome. Science 376, eabm1483 (2022).
Meyer, F. et al. Critical assessment of metagenome interpretation: the second round of challenges. Nat. Methods 19, 429–440 (2022).
Forsberg, K. J. et al. Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616 (2014).
Yin, X. L. et al. Global environmental resistome: distinction and connectivity across diverse habitats benchmarked by metagenomic analyses. Water Res. 235, 119875 (2023).
Calle, M. L. Statistical analysis of metagenomics data. Genomics Inform. 17, e6 (2019).
Diebold, P. J. et al. Clinically relevant antibiotic resistance genes are linked to a limited set of taxa within gut microbiome worldwide. Nat. Commun. 14, 7366 (2023).
Davidovich, C. et al. Occurrence of ‘under-the-radar’ antibiotic resistance in anthropogenically affected produce. ISME J. 19, wrae261 (2025).
Scott, T. M., Rose, J. B., Jenkins, T. M., Farrah, S. R. & Lukasik, J. Microbial source tracking: current methodology and future directions. Appl. Environ. Microb. 68, 5796–5803 (2002).
Bansal, M. S., Banay, G., Harlow, T. J., Gogarten, J. P. & Shamir, R. Systematic inference of highways of horizontal gene transfer in prokaryotes. Bioinformatics 29, 571–579 (2013).
Djordjevic, S. P. et al. Genomic surveillance for antimicrobial resistance—a One Health perspective. Nat. Rev. Genet. 25, 142–157 (2024).
Medvecky, M. et al. Interspecies transmission of CMY-2-producing sequence type 963 isolates between humans and gulls in Australia. mSphere 7, e0023822 (2022).
D’Costa, V. M., McGrann, K. M., Hughes, D. W. & Wright, G. D. Sampling the antibiotic resistome. Science 311, 374–377 (2006).
Wu, Y. et al. Wastewater treatment plant effluents exert different impacts on antibiotic resistome in water and sediment of the receiving river: metagenomic analysis and risk assessment. J. Hazard. Mater. 460, 132528 (2023).
Qian, X. et al. Long-read sequencing revealed cooccurrence, host range and potential mobility of antibiotic resistome in cow feces. Proc. Natl Acad. Sci. USA 118, e2024464118 (2021).
Wu, Z. et al. Nanopore-based long-read metagenomics uncover the resistome intrusion by antibiotic resistant bacteria from treated wastewater in receiving water body. Water Res. 226, 119282 (2022).
Che, Y. et al. High-resolution genomic surveillance elucidates a multilayered hierarchical transfer of resistance between WWTP- and human/animal-associated bacteria. Microbiome 10, 16 (2022).
Baron, S. A., Diene, S. M. & Rolain, J.-M. Human microbiomes and antibiotic resistance. Hum. Microbiome J. 10, 43–52 (2018).
Maestre-Carballa, L., Navarro-Lopez, V. & Martinez-Garcia, M. A resistome roadmap: from the human body to pristine environments. Front. Microbiol. 13, 858831 (2022).
Lee, K. et al. Population-level impacts of antibiotic usage on the human gut microbiome. Nat. Commun. 14, 1191 (2023).
Gacesa, R. et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 604, 732–739 (2022).
Karkman, A., Pärnänen, K. & Larsson, D. G. J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 10, 80 (2019).
Zhang, Z. et al. Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 13, 1553 (2022).
Christou, A. et al. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: the knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes—a review. Water Res. 123, 448–467 (2017).
Zhao, Q. & Liu, Y. Is anaerobic digestion a reliable barrier for deactivation of pathogens in biosludge? Sci. Total Environ. 668, 893–902 (2019).
Luo, Y. et al. Characteristics of wild bird resistomes and dissemination of antibiotic resistance genes in interconnected bird-habitat systems revealed by similarity of blaTEM polymorphic sequences. Environ. Sci. Technol. 56, 15084–15095 (2022).
Akter, S. et al. Detection of antibiotic-resistant bacteria and their resistance genes from houseflies. Vet. World 13, 266–274 (2020).
Zurek, L. & Ghosh, A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl. Environ. Microb. 80, 3562–3567 (2014).
Bogri, A. et al. Transmission of antimicrobial resistance in the gut microbiome of gregarious cockroaches: the importance of interaction between antibiotic exposed and non-exposed populations. mSystems 9, e0101823 (2024).
Aarestrup, F. M. The livestock reservoir for antimicrobial resistance: a personal view on changing patterns of risks, effects of interventions and the way forward. Philos. Trans. R. Soc. B Biol. Sci. 370, 20140085 (2015).
Tiseo, K., Huber, L., Gilbert, M., Robinson, T. P. & Van Boeckel, T. P. Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics 9, 918 (2020).
Dewulf, J. et al. Antibiotic use in European pig production: less is more. Antibiotics 11, 1493 (2022).
Muloi, D. M. et al. Population genomics of Escherichia coli in livestock-keeping households across a rapidly developing urban landscape. Nat. Microbiol. 7, 581–589 (2022).
Castro-Vargas, R. E., Herrera-Sanchez, M. P., Rodriguez-Hernandez, R. & Rondon-Barragan, I. S. Antibiotic resistance in Salmonella spp. isolated from poultry: a global overview. Vet. World 13, 2070–2084 (2020).
Zhang, Y. et al. Impacts of farmland application of antibiotic-contaminated manures on the occurrence of antibiotic residues and antibiotic resistance genes in soil: a meta-analysis study. Chemosphere 300, 134529 (2022).
Wang, F. et al. The overlap of soil and vegetable microbes drives the transfer of antibiotic resistance genes from manure-amended soil to vegetables. Sci. Total Environ. 828, 154463 (2022).
Schar, D. et al. Twenty-year trends in antimicrobial resistance from aquaculture and fisheries in Asia. Nat. Commun. 12, 5384 (2021).
Aarestrup, F. M. & Woolhouse, M. E. J. Using sewage for surveillance of antimicrobial resistance. Science 367, 630–632 (2020).
Beltran de Heredia, I. et al. Spatio-seasonal patterns of the impact of wastewater treatment plant effluents on antibiotic resistance in river sediments. Environ. Pollut. 319, 120883 (2023).
Wang, J., Chu, L., Wojnarovits, L. & Takacs, E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: an overview. Sci. Total Environ. 744, 140997 (2020).
Van Goethem, M. W. et al. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6, 40 (2018).
Qian, X. et al. Metagenomic analysis reveals the shared and distinct features of the soil resistome across tundra, temperate prairie and tropical ecosystems. Microbiome 9, 108 (2021).
Delgado-Baquerizo, M. et al. The global distribution and environmental drivers of the soil antibiotic resistome. Microbiome 10, 219 (2022).
Zheng, D. S. et al. Global biogeography and projection of soil antibiotic resistance genes. Sci. Adv. 8, eabq8015 (2022).
Xiao, R. et al. Antibiotic resistance in soil-plant systems: a review of the source, dissemination, influence factors and potential exposure risks. Sci. Total Environ. 869, 161855 (2023).
Han, Z. et al. Three-year consecutive field application of erythromycin fermentation residue following hydrothermal treatment: cumulative effect on soil antibiotic resistance genes. Engineering 15, 78–88 (2022).
McManus, P. S., Stockwell, V. O., Sundin, G. W. & Jones, A. L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40, 443–465 (2002).
Lorenzini, G. & Nali, C. Plant protection, the Cinderella of the one health strategy? One Health 20, 101080 (2025).
Miller, S. A., Ferreira, J. P. & LeJeune, J. T. Antimicrobial use and resistance in plant agriculture: a One Health perspective. Agriculture 12, 289 (2022).
Chen, P., Yu, K. & He, Y. The dynamics and transmission of antibiotic resistance associated with plant microbiomes. Environ. Int. 176, 107986 (2023).
van Rhijn, N. et al. Aspergillus fumigatus strains that evolve resistance to the agrochemical fungicide ipflufenoquin in vitro are also resistant to olorofim. Nat. Microbiol. 9, 29–34 (2024).
Ghaly, T. M., Chow, L., Asher, A. J., Waldron, L. S. & Gillings, M. R. Evolution of class 1 integrons: mobilization and dispersal via food-borne bacteria. PLoS ONE 12, e0179169 (2017).
Yin, Y., Zhu, D., Yang, G., Su, J. Q. & Duan, G. L. Diverse antibiotic resistance genes and potential pathogens inhabit in the phyllosphere of fresh vegetables. Sci. Total Environ. 815, 152851 (2022).
Reid, C. J., Blau, K., Jechalke, S., Smalla, K. & Djordjevic, S. P. Whole genome sequencing of Escherichia coli from store-bought produce. Front. Microbiol. 10, 3050 (2020).
Zhou, Z. C. et al. Association between particulate matter (PM) air pollution and clinical antibiotic resistance: a global analysis. Lancet Planet. Health 7, E649–E659 (2023).
Zhu, G. B. et al. Air pollution could drive global dissemination of antibiotic resistance genes. ISME J. 15, 270–281 (2021).
Bai, H. et al. Spread of airborne antibiotic resistance from animal farms to the environment: dispersal pattern and exposure risk. Environ. Int. 158, 106927 (2022).
Xie, J., Jin, L., Wu, D., Pruden, A. & Li, X. Inhalable antibiotic resistome from wastewater treatment plants to urban areas: bacterial hosts, dissemination risks and source contributions. Environ. Sci. Technol. 56, 7040–7051 (2022).
Li, L. et al. Municipal solid waste treatment system increases ambient airborne bacteria and antibiotic resistance genes. Environ. Sci. Technol. 54, 3900–3908 (2020).
Wu, D. et al. Inhalable antibiotic resistomes emitted from hospitals: metagenomic insights into bacterial hosts, clinical relevance and environmental risks. Microbiome 10, 19 (2022).
Kormos, D., Lin, K. S., Pruden, A. & Marr, L. C. Critical review of antibiotic resistance genes in the atmosphere. Environ. Sci. Process. Impacts 24, 870–883 (2022).
New, F. N. & Brito, I. L. What is metagenomics teaching us, and what is missed? Annu. Rev. Microbiol. 74, 117–135 (2020).
Forster, S. C. et al. Strain-level characterization of broad host range mobile genetic elements transferring antibiotic resistance from the human microbiome. Nat. Commun. 13, 1445 (2022).
Keenum, I. et al. A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater. Crit. Rev. Environ. Sci. Technol. 52, 4395–4419 (2022).
Ko, K. K. K., Chng, K. R. & Nagarajan, N. Metagenomics-enabled microbial surveillance. Nat. Microbiol. 7, 486–496 (2022).
Hendriksen, R. S. et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 10, 1124 (2019).
Risely, A. et al. Host-plasmid network structure in wastewater is linked to antimicrobial resistance genes. Nat. Commun. 15, 555 (2024).
Chen, X., Yin, X. L., Xu, X. Q. & Zhang, T. Species-resolved profiling of antibiotic resistance genes in complex metagenomes through long-read overlapping with Argo. Nat. Commun. 16, 1744 (2025).
Abramova, A., Berendonk, T. U. & Bengtsson-Palme, J. A global baseline for qPCR-determined antimicrobial resistance gene prevalence across environments. Environ. Int. 178, 108084 (2023).
Rhodes, J. et al. Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment. Nat. Microbiol. 7, 663–674 (2022).
Ferreira, C., Otani, S., Aarestrup, F. M. & Manaia, C. M. Quantitative PCR versus metagenomics for monitoring antibiotic resistance genes: balancing high sensitivity and broad coverage. FEMS Microbes 4, xtad008 (2023).
Carr, A., Diener, C., Baliga, N. S. & Gibbons, S. M. Use and abuse of correlation analyses in microbial ecology. ISME J. 13, 2647–2655 (2019).
Roy Chowdhury, P. et al. Phylogenomic analysis of a global collection of Escherichia coli ST38: evidence of interspecies and environmental transmission? mSystems 8, e0123622 (2023).
Kneis, D. et al. Trimethoprim resistance in surface and wastewater is mediated by contrasting variants of the gene. ISME J. 17, 1455–1466 (2023).
Gatica, J., Jurkevitch, E. & Cytryn, E. Comparative metagenomics and network analyses provide novel insights into the scope and distribution of beta-lactamase homologs in the environment. Front. Microbiol. 10, 146 (2019).
WHO. WHO Integrated Global Surveillance on ESBL-producing E. coli Using a ‘One Health’ Approach: Implementation and Opportunities (World Health Organization, 2021); https://www.who.int/publications/i/item/9789240021402
Watt, A. E. et al. Parameters for one health genomic surveillance of Escherichia coli from Australia. Nat. Commun. 16, 17 (2025).
Xu, X. et al. Ecological connectivity of genomic markers of antimicrobial resistance in Escherichia coli in Hong Kong. Nat. Commun. 16, 7319 (2025).
Muloi, D. M. et al. Exploiting genomics for antimicrobial resistance surveillance at One Health interfaces. Lancet Microbe 4, e1056–e1062 (2023).
Anjum, M. F. et al. The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opin. Microbiol. 64, 152–158 (2021).
Holcomb, D. A. & Stewart, J. R. Microbial indicators of fecal pollution: recent progress and challenges in assessing water quality. Curr. Environ. Health Rep. 7, 311–324 (2020).
Horesh, G. et al. A comprehensive and high-quality collection of genomes and their genes. Microb. Genom. 7, 000499 (2021).
Karp, Peter et al. The EcoCyc database. EcoSal Plus 11, eesp-0002 (2023).
Shi, X. et al. Microbial risk assessment across multiple environments based on metagenomic absolute quantification with cellular internal standards. Nat. Water 3, 473–485 (2025).
Yang, Y. et al. Establishing reference material for the quest towards standardization in environmental microbial metagenomic studies. Water Res. 245, 120641 (2023).
Zhu, Y. G. et al. Microbial mass movements. Science 357, 1099–1100 (2017).
WHO. UN General Assembly High-Level Meeting on Antimicrobial Resistance 2024 (World Health Organization, 2024); https://www.who.int/news-room/events/detail/2024/09/26/default-calendar/un-general-assembly-high-level-meeting-on-antimicrobial-resistance-2024
Lacroix, M. Z. et al. Residues of veterinary antibiotics in manures from pig and chicken farms in a context of antimicrobial use reduction by implementation of health and welfare plans. Environ. Res. 238, 117242 (2023).
Van Epps, A. & Blaney, L. Antibiotic residues in animal waste: occurrence and degradation in conventional agricultural waste management practices. Curr. Pollut. Rep. 2, 135–155 (2016).
Chen, Z. Y. et al. Unraveling the influence of human fecal pollution on antibiotic resistance gene levels in different receiving water bodies using crAssphage indicator gene. J. Hazard. Mater. 442, 130005 (2023).
Acknowledgements
This work is supported by the General Research Fund (17202522), the National Natural Science Foundation of China (22193062) and the Theme-based Research Scheme of Research Grants Council of Hong Kong (T21–705/20-N).
Author information
Authors and Affiliations
Contributions
T.Z., L.L., B.L., X.Y. and E.T. conceptualized the study. T.Z. provided supervision and guided the overall direction. L.L. designed the figures and wrote the manuscript, with input from all authors. Y.X., Y.Y. and X.X. contributed to visualization. M.R.G., W.G., M.J.B., C.M.M., D.G., K.S., S.P.D., A.P., P.V., E.C., E.D., N.A., G.C., D.F.-K., F.W. and T.U.B. contributed to discussion of the content, and writing and editing of the manuscript. P.J.J.A., M.v.L., P.H.N., R.H., B.F.S., D.F., T.T.-Y.L., K.M.Y.L., F.X., X.Z., J.G., H.S., G.D.W., J.M., C.B., R.C.P., S.Z.A., C.-J.C., G.Y., Y.L., Y.W., J.S., Y.Z., M.Y., X.L., B.H., L.Z., Y.W., S.T., B.K. and Y.-G.Z. commented on the paper. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Water thanks Davida Smyth and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, L., Li, B., Yin, X. et al. Assessing antimicrobial resistance connectivity across One Health sectors. Nat Water 3, 1100–1113 (2025). https://doi.org/10.1038/s44221-025-00514-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44221-025-00514-8
This article is cited by
-
Towards One Health action for addressing antimicrobial resistance in the age of polycrisis
Nature Sustainability (2026)