Figure 1
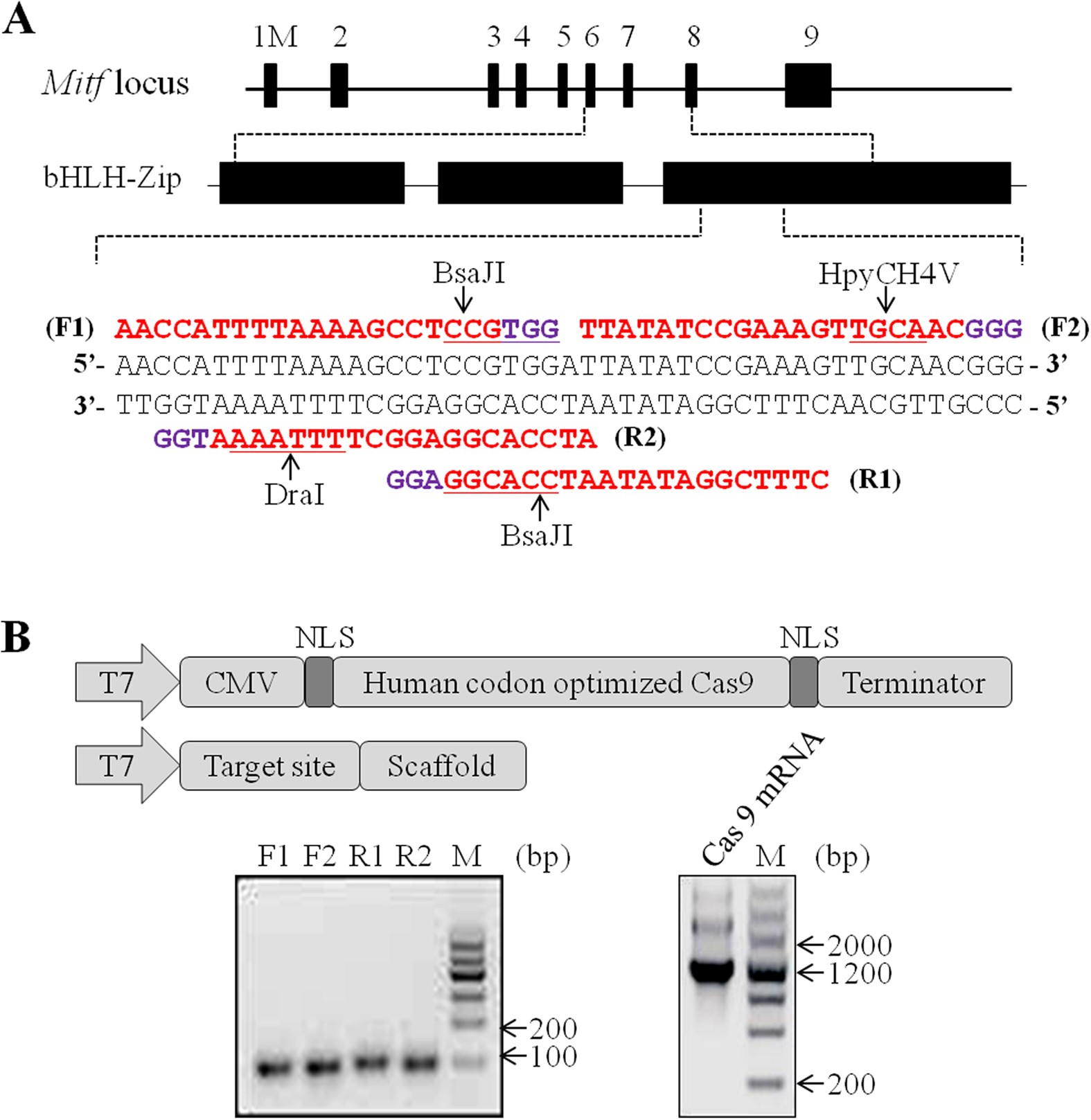
Design and construction of CRISPR.
(A) Schematic of the Cas9/sgRNA-targeting sites in pig mitfloci. Exons are shown as black boxes. The sgRNA-targeting sequence is labeled in red and the protospacer-adjacent motif (PAM) sequence is labeled in purple. The different sgRNAs are marked as F1, F2, R1 and R2, respectively. The restriction sites at the target regions are underlined. Restriction enzymes used for RFLP analysis are shown. (B) Top: schematic diagram of the templates for in vitro transcription used to generate the Cas9 mRNA and sgRNA. Bottom: assessing the quality of sgRNAs and Cas9 mRNA by gel electrophoresis. Electrophoresis of sgRNAs yields a single band approximately 100 bp. Electrophoresis of Cas9 mRNA yields 2–3 bands due to persistent secondary structure. The quality of mRNA is good if discrete bands are visible.
