Abstract
The ErbB-2 interacting protein receptor-associated late transducer (RALT) was previously identified as a feedback inhibitor of ErbB-2 mitogenic signals. We now report that RALT binds to ligand-activated epidermal growth factor receptor (EGFR), ErbB-4 and ErbB-2.ErbB-3 dimers. When ectopically expressed in 32D cells reconstituted with the above ErbB receptor tyrosine kinases (RTKs) RALT behaved as a pan-ErbB inhibitor. Importantly, when tested in either cell proliferation assays or biochemical experiments measuring activation of ERK and AKT, RALT affected the signalling activity of distinct ErbB dimers with different relative potencies. RALT ΔEBR, a mutant unable to bind to ErbB RTKs, did not inhibit ErbB-dependent activation of ERK and AKT, consistent with RALT exerting its suppressive activity towards these pathways at a receptor-proximal level. Remarkably, RALT ΔEBR retained the ability to suppress largely the proliferative activity of ErbB-2.ErbB-3 dimers over a wide range of ligand concentrations, indicating that RALT can intercept ErbB-2.ErbB-3 mitogenic signals also at a receptor-distal level. A suppressive function of RALT ΔEBR towards the mitogenic activity of EGFR and ErbB-4 was detected at low levels of receptor occupancy, but was completely overcome by saturating concentrations of ligand. We propose that quantitative and qualitative aspects of RALT signalling concur in defining identity, strength and duration of signals generated by the ErbB network.
Similar content being viewed by others
Introduction
The family of ErbB receptor tyrosine kinases (RTKs) consists of four members: ErbB-1 (also referred to as epidermal growth factor receptor, EGFR) through ErbB-4. In the human genome, a dozen 10 genes encode ErbB ligands possessing distinct receptor specificity. Regardless of the latter, all ErbB ligands trigger cognate receptors via a common mechanism entailing the assembly of homo- and heterodimeric receptor combinations (reviewed in Olayioye et al., 2000; Yarden and Sliwkowski, 2001). The ensuing combinatorial repertoire of ligand-driven receptor dimers provides the family of ErbB RTKs with a great deal of signalling versatility and, consequently, the ability to govern a wide array of cellular programs. This is best exemplified by genetic studies, which have shown that defined combinations of ErbB receptors play nonredundant roles during development and in postnatal life. In human pathology, this is mirrored by the selective derangement of EGFR and ErbB-2 activity during the pathogenesis of a number of tumors (reviewed in Olayioye et al., 2000; Yarden and Sliwkowski, 2001).
ErbB-1 and ErbB-4 share canonical features of RTKs and can generate downstream signals upon ligand-induced homodimerization. In contrast, ErbB-2 and ErbB-3 are peculiar in that ligand-driven lateral interactions with other ErbB RTKs represent a sine qua non for initiation of their signalling activity (reviewed in Olayioye et al., 2000; Yarden and Sliwkowski, 2001). ErbB-3 is able to bind ligands, such as neuregulins (NRGs), directly (Carraway III et al., 1994, 1997), but is devoid of intrinsic catalytic activity (Guy et al., 1994). Therefore, ErbB-3 acquires signalling capacity only in the context of heterodimers with catalytically competent ErbB RTKs (Pinkas-Kramarski et al., 1996). Conversely, ErbB-2 is unable to bind ligands directly, but behaves as a coreceptor for several ligands when complexed with other ErbB RTKs (Graus-Porta et al., 1995, 1997; Riese et al., 1995; Karunagaran et al., 1996; Riese and Stern, 1998). In fact, ErbB-2 appears to be the favorite partner of all other ErbB receptors and emerges as the hierarchically dominant element in the assembly of ErbB heterodimers (Graus-Porta et al., 1997). Of particular relevance is the ErbB-2/ErbB-3 combination that reconstitutes a high-affinity receptor for NRGs endowed with potent oncogenic activity (Alimandi et al., 1995; Wallasch et al., 1995).
Individual ErbB receptors may couple selectively to some signalling pathways: for example, only EGFR is able to activate STAT1 and STAT3 (Olayioye et al., 1999). Combinatorial dimerization among ErbB RTKs generates additional signalling potential via integration of the signalling competence of individual protomers as well as the generation of novel signalling profiles (Olayioye et al., 1998). Finally, ErbB RTKs share some signalling pathways, such as those leading to Ras–ERK and PI-3K–AKT activation. Importantly, distinct ErbB dimers may activate these pathways with diverse profiles of intensity and duration (Olayioye et al., 2000 and references therein), which may be themselves critical determinants of signal specificity (Marshall, 1995; Ghiglione et al., 1999). This raises the question of how strength and duration of ErbB signals are proofread in order to (a) allow the correct execution of cellular programs governed by precise thresholds of receptor signals and (b) restrain the oncogenic potential of ErbB activity.
It is increasingly appreciated that negative signalling is instrumental in defining quantitative aspects of ErbB outputs. Coupling to internalization/degradation pathways is thought to set thresholds of activity of ErbB RTKs at the inception of receptor triggering (reviewed in Waterman and Yarden, 2001). Much less studied are the mechanisms that gauge and eventually extinguish ErbB signals as they accumulate over time because of sustained receptor activity. Genetic studies in flies indicate that expression of Drosophila EGFR (DER) feedback inhibitors such as Kekkon1, Argos and Sprouty is controlled transcriptionally by DER via the Ras-ERK pathway (Perrimon and McMahon, 1999). Importantly, negative feedback regulation of DER signalling is critical for ensuring that DER activity be confined within appropriate spatial and temporal boundaries (Perrimon and McMahon, 1999; Freeman, 2000). Thus, it appears that the levels of expression of Aos, Spry and Kek1 are a means for the cell to gauge the accumulation of receptor signals, while their activity is uniquely suited to proofread, tune and extinguish DER activity. Consequently, it has been proposed that negative feedback loops provide DER signalling with an essential element of stability, which prevents perturbation of DER function and allows for accurate reproduction of signals responsible for complex developmental patterns (Freeman, 2000).
Receptor-associated late transducer (RALT) has been recently identified as a feedback inhibitor of mitogenic signals propagated by ErbB-2 (Fiorentino et al., 2000) and EGFR (Hackel et al., 2001). RALT expression is activated by EGF, TGF-α and NRG-1 in fibroblasty and epithelial cells (Fiorentino et al., 2000), via a mechanism entailing transcriptional activation of the Ralt gene by the Ras–Raf–ERK pathway (Fiorini et al., 2002). Constitutive overexpression of RALT markedly inhibits the mitogenic and transforming activity of EGFR (Hackel et al., 2001) and ErbB-2 (Fiorentino et al., 2000). Conversely, neutralization of endogenous RALT function by microinjection of anti-RALT antibodies enhances the mitogenic activity of an EGFR/ErbB-2 chimeric receptor (Fiorini et al., 2002). These data indicate that physiological levels of RALT are involved in the suppression of ErbB-2 signals and imply that RALT function may in fact play a necessary role in negative regulation of ErbB-2 activity. Owing to its ability to complex with ErbB-2- and SH3-containing proteins, RALT has been proposed to function as a receptor-proximal inhibitor, which links ErbB-2 to effectors containing SH3 domains (Fiorentino et al., 2000). However, the significance of the RALT/ErbB-2 physical interaction in the context of RALT-mediated inhibition of ErbB-2 signalling has not been formally addressed, nor is it known whether and to what extent RALT is able to antagonize mitogenic signals propagated by the entire ErbB signalling network. The latter is an especially relevant issue, given the high degree of signal integration among ErbB RTKs.
Here, we report that RALT functions as a pan-ErbB inhibitor. Remarkably, the potency of RALT suppressive activity impacts differentially on individual ErbB receptors. We propose that RALT activity adds a novel layer of regulation to the specification of signal output by the ErbB network.
Results
RALT is not a promiscuous RTK inhibitor
Despite its suppressive activity towards mitogenic and transforming signals triggered by ErbB-2 and EGFR, ectopically expressed RALT altered neither mitogenic responses of murine fibroblasts to serum nor cell transformation induced by v-ras and v-src (Fiorentino et al., 2000; Hackel et al., 2001). The issue of RALT specificity was further addressed in the experiments shown in Figure 1. NIH-EGFR/ErbB-2 cells were infected with either Pinco or Pinco–RALT retroviral stocks. RALT expression was induced in Pinco cells by EGF (Figure 1a) as well as by bFGF and PDGF (data not shown, see also Fiorini et al., 2002) with comparable kinetics. Pinco–RALT cells showed constitutive expression of ectopic RALT at levels 3–4-fold higher than the endogenous species (Figure 1a). Whereas ErbB-2-dependent mitogenic signals were clearly suppressed by RALT overexpression (42–52% reduction over a 0.3–5 ng ml EGF concentration range), proliferative responses to PDGF and bFGF were comparable in Pinco and Pinco–RALT cells over a wide range of ligand concentration (Figure 1b); also proliferative responses to PMA were unaffected by RALT overexpression (data not shown). These results indicate that RALT is unlikely to behave as a pan-RTK inhibitor.
Biological analysis of RALT signalling in mitogenic programs activated by PDGF and bFGF. NIH-EGFR/ErbB-2 cells were seeded in 24-well plates and infected with either Pinco or Pinco–RALT retrovirus stocks. Infection efficiency, as monitored by FACS assessment of GFP-positive cells was >95%. (a) Following infection, cells were made quiescent by 24 h serum deprivation and subsequently challenged with 5 ng/ml EGF for the indicated time. Cell lysates were subjected to immunoblot analysis with either the S1 antiserum that recognizes mouse, rat and human RALT species or MoAb 19C5/4 specific for the ectopically expressed rat RALT species. (b) Following infection, cells were switched to either serum-free medium (SFM) or SFM containing escalating concentrations of EGF (0.3, 1 and 5 ng/ml), bFGF (0.3, 1 and 5 ng/ml) or PDGF (0.2, 2 and 10 ng/ml). Assays were performed in quadruplicate wells. After 44 h, cultures were labelled for 4 h with 1 μCi/ml [3H]methyl-thymidine and cell proliferation measured as fold increase of TCA-precipitated radioactivity in growth factor-treated wells versus control cultures. The s.d. was <15%. Data are representative of one out of three experiments that gave similar results. White bars indicate Pinco cells, gray bars indicate Pinco–RALT cells
RALT binds in vitro to all members of the ErbB family
Based on the above conclusions, we focused our attention on the function of RALT within the ErbB signalling network. Thus, we asked whether RALT could enter in a complex with ErbB RTKs other than ErbB-2.
Previous studies have shown that the ErbB-2-binding activity of RALT is confined to the RALT COOH terminus (residues 262–459); within this 197 amino-acid-long stretch, the sequence comprised between positions 282 and 396 (GST-RALT cl.52) was sufficient to bind ErbB-2 in GST pull down assays and in yeast two-hybrid experiments (Fiorentino et al., 2000) (see also Figures 2a and 3). GST-RALT cl.52 was able to precipitate ErbB-2 and ErbB-3 from lysates of NIH-E2.E3 transfectants; ErbB-3 could be precipitated only from lysates of NRG-1-stimulated cells, whereas ErbB-2 bound to GST-RALT cl.52 irrespective of NRG-1 stimulation (Figure 2a). In the absence of ErbB-2 coexpression, ErbB-3 was not precipitated by GST-RALT cl.52, even under conditions of optimal stimulation with NRG-1. On the contrary, ErbB-2 bound efficiently to GST-RALT also in the absence of ectopically expressed ErbB-3. The ligand-independent binding of ErbB-2 to GST-RALT cl.52 is most likely a consequence of the constitutive activation of ErbB-2 in NIH 3T3 transfectants, as reported previously (Lonardo et al., 1990) and demonstrated herein by the anti-P-Tyr immunoblot shown in Figure 2a. Binding to GST-RALT cl.52 was also observed with EGFR (Figure 2b) and ErbB-4 (Figure 2c) solubilized from NIH-E1 and NIH-E4 transfectants, respectively. Binding of ErbB-4 to GST–RALT cl.52 was unaffected by EGF stimulation but enhanced by NRG-1 stimulation. Conversely, EGF but not NRG-1 stimulation enhanced binding of EGFR to GST–RALT cl.52.
In vitro analysis of RALT interaction with ErbB RTKs. (a) Recombinant GST-RALT cl.52 fusion protein (referred to as GST–RALT) spanning positions 282–396 of RALT was purified onto glutathione–sepharose beads and used in pull-down assays against ErbB receptors solubilized from the indicated NIH 3T3 transfectants. Lysates of parental NIH 3T3 were used as control. Where indicated, cells were treated with 20 ng/ml recombinant NRG-1β for 5 min prior to lysis. Protein complexes were subjected to immunoblot analysis with the indicated antibodies. The bottom portion of the gel was stained with Coomassie blue to control for equal input of GST–RALT. Reference lysates (immunoblotted with anti-ErbB-3 and anti-P-Tyr antibodies) corresponded to 5% of input lysate in GST pull-down experiments. Control experiments (data not shown, see also Figure 3b) indicated that GST alone did not bind to ErbB RTKs. (b and c) NIH-EGFR (b) and NIH-ErbB-4 (c) transfectants were serum starved for 16 h and lysed either without further treatment or after stimulation for 5 min with 20 ng/ml of the indicated ligands. Lysates were subjected to pull-down assays with GST–RALT and immunoblotted with anti-EGFR (b) and anti-ErbB-4 antibodies (c). Reference lysates corresponded to 5% of input lysate in GST pull-down experiments. Input GST–RALT was visualized by Coomassie stain
Mapping of the minimal EBR in the RALT protein. (a, top) Schematic outline of domain organization of RALT and RALT ΔEBR mutant. NLS indicates a putative nuclear localization signal. (Bottom) Schematic representation of RALT fragments used as GST fusion products in pull-down assays reported in panel b. The ability of each recombinant polypeptide to interact in vitro with the ErbB-2 protein is also indicated. Note that RALT fragments are drawn to a scale larger than the one used to draw RALT and RALT ΔEBR in the top portion of the panel. (b) GST fusion proteins (5 μg for each pull-down experiment) spanning the indicated portions of the RALT protein were produced in E. coli, purified onto glutathione–agarose beads and assayed for their ability to capture ErbB-2 protein solubilized from NIH-ErbB-2 transfectants. Protein complexes were analysed by immunoblot with anti-ErbB-2 antibodies. GST was used as control; reference lysate corresponded to 5% of input lysate in pull-down assays
Identification of the minimal ErbB-2-binding region of RALT
In order to define the minimal ErbB-2-binding region (EBR) of RALT, we generated a panel of GST–RALT fusion proteins (Figure 3a) and assayed their ability to precipitate ErbB-2 from lysates of NIH-ErbB-2 transfectants. Neither the NH2 (aa. 282–343) nor the COOH (aa. 335–396) halves of GST–RALT cl.52 were capable of binding to ErbB-2 (data not shown), whereas sequences comprised between positions 282 and 312 were dispensable for directing the interaction of GST–RALT cl.52 with ErbB-2 (Figure 3b). Further deletion analysis defined the sequence comprised between positions 323 and 372 of RALT as the minimal EBR (Figure 3b). We took advantage of a conveniently located StuI site in the Ralt cDNA sequence to generate a RALT mutant lacking residues 315–361 (RALT ΔEBR, Figure 3a), which could be expressed in various cell lines as efficiently as full-length RALT (see below).
Analysis of RALT interaction with ErbB receptors in living cells
We next asked whether RALT interacts with ErbB RTKs in living cells in an EBR-dependent fashion. To address these questions, we performed coimmunoprecipitation assays using lysates of either 293 or NIH 3T3 cells engineered to express ErbB RTKs along with either RALT or RALT ΔEBR (Figures 4 and 5). Neither RALT nor RALT ΔEBR had detrimental consequences on steady-state levels of coexpressed ErbB receptors (Figures 4 and 5). Importantly, the ΔEBR mutant was able to interact with Grb-2 as efficiently as RALT (Figure 4b). We infer that the ΔEBR mutation is unlikely to cause gross conformational alterations of the RALT protein, since the RALT/Grb-2 interaction requires that Grb-2 SH3 domains bind to Pro-rich sequences located in close proximity to the RALT EBR module (Fiorentino et al., 2000).
In vivo analysis of physical interaction between RALT and the ErbB-2.ErbB-3 NRG-1 receptor. (a) ErbB-2 and ErbB-3 were transiently expressed in 293 cells either alone or in combination. Where indicated, either RALT or RALT ΔEBR were coexpressed with ErbB RTKs. After 2 h serum starvation, cells were stimulated at 37°C with either carrier solution or 20 ng/ml NRG-1β before lysis. Equal amounts of each lysate were analysed by immunoblot with anti-ErbB-2 and anti-ErbB-3 antibodies to control for receptor expression. Lysates were subjected to immunoprecipitation with anti-RALT 19C5/4 monoclonal antibody followed by blot analysis with anti-RALT and anti-ErbB-3 antibodies. Filters were subsequently reprobed with anti-ErbB-2 antibodies. (b) The ΔEBR mutation does not affect the ability of RALT to interact with Grb-2. HA-tagged GRB-2 was expressed in 293 cells either alone or in combination with RALT or RALT ΔEBR. Equal amounts of each lysate and anti-RALT immunoprecipitates were immunoblotted with anti-HA tag 12CA/5 and anti-RALT 19C5/4 monoclonal antibodies
In vivo analysis of RALT interaction with EGFR and ErbB-4. (a) NIH 3T3 and NIH-EGFR cells were infected with recombinant retrovirus directing the expression of either RALT or RALT ΔEBR. Control cells were infected with empty virus. After 2 h serum starvation, cells were stimulated for 5 min with either carrier solution or EGF (10 ng/ml) prior to lysis. Equal amounts of each lysate were analysed by immunoblot with anti-receptor and anti-P-Tyr antibodies. Lysates were subjected to immunoprecipitation with anti-RALT 19C5/4 monoclonal antibody followed by blot analysis with anti-RALT and anti-receptor antibodies. (b) ErbB-4 was transiently expressed in 293 cells along with either RALT or RALT ΔEBR. At 48 h after transfection, cells were lysed after stimulation for 5 min with either carrier solution or 20 ng/ml NRG-1. Equal amounts of each lysate were analysed by immunoblot with anti-ErbB-4 and anti-P-Tyr antibodies. Anti-RALT 19C5/4 immunoprecipitates were resolved on SDS–PAGE and immunoblotted with anti-RALT and anti-ErbB-4 antibodies. (c) RALT was coexpressed in 293 cells with either wt EGFR or EGFR mutants depicted in the left portion of the panel, namely EGFR K721A (KD, i.e. kinase-defective) and EGFR ΔC, that lacks the COOH-terminal region. At 48 h after transfection, cells were stimulated for 5 min with either carrier solution or 10 ng/ml EGF. Equal amounts of each lysate were analysed by immunoblot with anti-RALT and anti-P-Tyr antibodies. Anti-EGFR immunoprecipitates were resolved on SDS–PAGE and immunoblotted with anti-RALT and anti-EGFR antibodies
RALT, but not RALT ΔEBR, was found in a complex with ErbB-2 in 293 cells, as revealed by probing anti-RALT immunoprecipitates with anti-ErbB-2 antibodies. This immunoreactivity was contingent on coexpression of RALT and ErbB-2 (Figure 4a). ErbB-3 could be coimmunoprecipitated with anti-RALT antibodies only if coexpressed with ErbB-2 and solubilized from cells stimulated with NRG-1. Coimmunoprecipitation of RALT and ErbB-3 required integrity of the EBR (Figure 4a). Thus, it appears that ErbB-3 coimmunoprecipitates with RALT only when recruited to ErbB-2.ErbB-3 dimers generated by NRG-1 stimulation. Interestingly, in E2.E3/RALT transfectants the amount of ErbB-2 immunoreactivity recovered in anti-RALT immunoprecipitates from NRG-stimulated cells was consistently lower than that obtained from unstimulated cells (Figure 4a). We infer that in unstimulated ErbB-2.ErbB-3 transfectants, RALT is likely to be complexed to activated ErbB-2 homodimers generated by ErbB-2 overexpression. NRG-1 stimulation drives redistribution of activated ErbB-2 molecules into complexes with ErbB-3, thus causing preferential association of RALT with ErbB-2.ErbB-3 heterodimers rather than with ErbB-2 homodimers.
EGFR (Figure 5a, c) and ErbB-4 (Figure 5b) were also able to associate with RALT. These interactions were largely dependent on ligand stimulation of the receptors and absolutely contingent on the presence of the EBR module in the RALT protein. A catalytically inactive EGFR mutant carrying the K-A mutation at position 721 was unable to recruit RALT in a physical complex both in anti-EGFR immunoprecipitations followed by anti-RALT immunoblot (Figure 5c), as well as in anti-RALT immunoprecipitations followed by anti-EGFR immunoblot (data not shown). Moreover, we did not detect RALT/EGFR K721A complexes even upon marked receptor overexpression, that is, under conditions causing constitutive, ligand-independent RALT/EGFR association (data not shown). In contrast, the deletion of the EGFR COOH tail, which contains all the major autophosphorylation sites of the receptor (ΔC mutant), did not affect RALT/EGFR complex formation (Figure 5c).
Collectively, the experiments described in Figures 3, 4 and 5 indicate that RALT interacts with different ErbB RTKs via the EBR module. Importantly, when comparing the amount of ErbB receptors precipitated by either GST–RALT or anti-RALT antibodies to the amount of ErbB receptors present in 5% of input lysate, we consistently observed that RALT interacts with EGFR, ErbB-4 and ErbB-2.ErbB-3 dimers with comparable efficiencies.
RALT is spatially controlled by its interaction with ligand-activated ErbB RTKs
EGF-dependent activation of an EGFR/ErbB-2 chimeric receptor leads to the redistribution of RALT from a cytosolic location to intracellular membranes (Fiorentino et al., 2000). The inability of the ΔEBR mutant to associate with ErbB RTKs allowed us to test whether physical interaction with ErbB receptors is implicated in spatial control of the RALT protein. MoAb 19C5/4 allows immunohistochemical detection of RALT and RALT ΔEBR with similar efficiency. Since Ab 19C5/4 does not crossreact with murine RALT (Fiorentino et al., 2000, see also Figure 1a), we could selectively image ectopic RALT proteins in NIH-EGFR and NIH-EGFR/ErbB-2 cells. Of note, the degree of RALT overexpression in these cells was modest (3–4-fold higher than that of the endogenous species, see Figure 1a for reference). Immunofluorescence imaging of RALT in quiescent cells indicated that both RALT (Figure 6e, m) and RALT ΔEBR (Figure 6g, o) were distributed in the cytosol. RALT relocated to a vesicular compartment following activation of EGFR for 15 min (Figure 6n) and EGFR/ErbB-2 for 15 min (not shown) or 60 min (Figure 6f), showing extensive colocalization with ErbB receptors (Figure 6j). Strikingly, ligand activation of neither EGFR nor EGFR/ErbB-2 caused relocation of RALT ΔEBR (Figure 6h, p). We conclude that RALT relocation induced by activation of ErbB RTKs requires that RALT forms a physical complex with ligand-bound ErbB RTKs. We also infer that the ΔEBR mutation does not abolish coimmunoprecipitation between RALT and ErbB receptors by simply causing a reduction of the affinity of RALT ΔEBR for ErbB RTKs.
Distinct spatial distribution of RALT and RALT ΔEBR in response to activation of ErbB receptors. Subconfluent cultures of serum-deprived NIH-EGFR/ErbB-2 (a–l) and NIH-EGFR (m–p) cells ectopically expressing either RALT or RALT ΔEBR were processed for immunocytochemistry before (a, e, i, c, g, k, m, o) or after stimulation with EGF (10 ng/ml) for 60 (b, f, j, d, h, l) or 15 min (n, p). NIH-EGFR/ErbB-2 cells (a–l) were double stained for ErbB-2 (a–d, green) and either RALT (e, f, red) or RALT ΔEBR (g, h, red). Upon EGF stimulation, RALT (f) relocates to vesicular perinuclear structures where it colocalizes with the EGFR/ErbB-2 chimera (b, merge in j), while RALT ΔEBR (h) does not show significant relocation and does not colocalizes with EGFR/ErbB-2 (d, merge in l). Also in EGF-treated NIH-EGFR cells (m–p), RALT (m, n) relocates from the cytosol to a vesicular compartment, whereas spatial distribution RALT ΔEBR (o, p) is not detectably altered by EGFR activation. Bars: 10 μm.
RALT inhibits ErbB signals leading to activation of ERKs and AKT
ErbB family members share the ability to trigger the Ras-ERK and the PI-3-AKT pathways (Yarden and Sliwkowski, 2001). Thus, we surmised that evaluation of ERK and AKT activities triggered by engagement of ErbB RTKs could provide a convenient biochemical readout for comparative analysis of RALT signalling to distinct ErbB dimers.
For these studies we used 32D cells reconstituted with defined ErbB RTKs. 32D.ErbB transfectants are relieved from IL-3 dependency if cultured in media containing ErbB ligands, thus providing a sensitive assay of the signalling activity of defined ErbB dimers (Alimandi et al., 1997). 32D.ErbB transfectants were further engineered to express either RALT or RALT ΔEBR. Of note, 32D cells do not express endogenous RALT protein, as indicated by immunoblot analysis of lysates prepared from 32D and 32D.E1 cells triggered with either IL-3 (growth medium) or EGF (Figure 7c). This was confirmed by Northern blot analysis (data not shown), in agreement with the absence of Mig-6 mRNA expression in cells of the hemopoietic lineage reported in the Genecard database (http://bioinfo.weizmann.ac.il/cards/). Thus, by expressing RALT proteins in 32D.ErbB transfectants we could (a) assess whether reconstitution of RALT expression is per se capable of restoring a pathway of negative signalling to ErbB RTKs and (b) study the activity of the ΔEBR mutant without the possible interference of endogenous RALT protein.
Analysis of expression of RALT, RALT ΔEBR and ErbB RTKs in 32D transfectants. (a) 32D transfectants expressing EGFR, ErbB-4 and the ErbB-2.ErbB-3 combination were supertransfected with Pinco, Pinco–RALT and Pinco–RALT ΔEBR expression vectors. Stable Pinco, RALT and RALT ΔEBR derivatives were established by puromycin selection. Lysates prepared from each of the transfectants were immunoblotted with antibodies specific for each of the ErbB family members. Filters were also probed with anti-SHC antibodies to control for equal loading of lysates. (b) lysates from 32D transfectants described in (a) were analysed simultaneously for RALT expression by immunoblot in order to compare relative levels of RALT expression. Filters were stripped and reprobed with anti-SHC antibodies to control for sample loading. (c) 32D and 32D.E1 cells were deprived of IL-3 for 2 h and then stimulated for the indicated time with either IL-3 (growth medium) or EGF (10 ng/ml). Equal amounts of lysates were immunoblotted with the indicated antibodies. A lysate from 32D.E1 RALT cells was used as positive control for RALT immunodetection
Stable RALT and RALT ΔEBR derivatives of 32D.E1, 32D.E2.E3 and 32D.E4 cells were generated by transfection with Pinco–RALT and Pinco–RALT ΔEBR vectors, respectively. Control cells were drug- selected after transfection with the backbone Pinco vector. Neither RALT nor RALT ΔEBR affected the steady-state levels of ErbB RTKs in 32D transfectants (Figure 7a). For each of the 32D.ErbB transfectants mentioned above, we selected cell populations expressing comparable amounts of RALT and RALT ΔEBR. Levels of RALT proteins were similar in 32D.E2.E3 and 32D.E4 cells. When compared to 32D.E4 and 32D.E2.E3 cells, 32D.E1 transfectants contained lower levels of ectopic RALT and RALT ΔEBR (Figure 7b).
In 32D.E1 Pinco cells EGF stimulation (3 ng/ml) led to sustained activation of ERKs throughout the 6 h time interval of the experiment. AKT was also activated robustly in the initial 60 min of EGF stimulation, after which time anti-phospho-AKT reactivity declined (Figure 8a). In EGF-treated 32D.E1 RALT ΔEBR cells strength and duration of ERK and AKT activation were quite similar to those observed in Pinco controls. In contrast, expression of RALT in 32D.E1 cells led to premature termination of ERK and AKT activation, with no reactivity to anti-P-ERK and anti-P-AKT being observed past the 1 h and 15 min time point, respectively (Figure 8a). These differences were not accounted for by changes in either EGFR expression or EGFR phosphorylation on tyrosine residues (Figure 8a).
Interaction of RALT with ErbB receptors affects strength and duration of ERK and AKT activation. Pinco, RALT and RALT ΔEBR derivatives of 32D.E1 (a), 32D.E2.E3 (b) and 32D.E4 (c) transfectants described in Figure 7 were deprived of IL-3 for 2 h. Following stimulation with 3 ng/ml EGF (a) or 10 ng/ml NRG-1β (b, c) for the indicated time, cells were harvested and lysed in Laemmli buffer. Lysates were analysed by immunoblot with the indicated antibodies. (a–c) Show a representative experiment of at least three independent assays for E1, E2.E3 and E4 transfectants
Stimulation of Pinco and RALT ΔEBR derivatives of 32D.E2.E3 cells with 10 ng/ml NRG-1 generated rather similar kinetics of activation of AKT and ERKs (Figure 8b). While the profile of ERK activity in 32D.E1 Pinco and 32D.E2.E3 Pinco transfectants were comparable, activated ErbB-2.ErbB-3 dimers generated a more sustained AKT signal (compare Figure 8a, b). This observation is consistent with ErbB-3 providing powerful activation of PI-3K once transphosphorylated by catalytically competent ErbB RTKs. Expression of ectopic RALT in 32D.E2.E3 cells caused premature extinction of both ERK and AKT activity (Figure 8b). ERK activity in E2.E3 RALT cells returned to background levels past the 3 h time point, that is, later than what we observed in E1 RALT transfectants (Figure 8a). AKT activity was not detectable past the 3 h time point in RALT cells, whereas it was still high after 9 h of NRG-1 stimulation in both E2.E3 Pinco and E2.E3 RALT ΔEBR cells. Neither RALT nor RALT ΔEBR altered significantly the profile of expression and activation of ErbB-2 and ErbB-3 (Figure 8b).
At variance with what observed in E1 and E2.E3 transfectants, expression of RALT in 32D.E4 cells led to a severe inhibition of NRG-1-dependent ERK activity since the inception of receptor triggering and throughout the 6 h time frame of the experiment (Figure 8c). Pronounced attenuation of anti-P-AKT reactivity in 32D.E4 RALT cell lysates was also observed at the earliest time point of NRG-1 stimulation, with P-AKT levels being reduced by 80% past the 60 min time point (Figure 8c). These drastic effects of RALT activity on ErbB-4 signalling could not be accounted for by obvious alterations in levels of ErbB-4 expression and tyrosine phosphorylation (Figure 8c). The ΔEBR mutation caused RALT to lose its ability to suppress ErbB-4-dependent activation of ERKs and AKT (Figure 8c). In fact, the activity of ERKs and AKT was reproducibly enhanced by the expression of RALT ΔEBR, raising the possibility that the ΔEBR mutant exerts a dominant-negative function (see the Discussion section).
Collectively, the data presented in Figure 8 indicate that genetic reconstitution of RALT expression in 32D.E1, 32D.E4 and 32D.E2.E3 cells is sufficient to suppress ErbB-dependent activation of ERKs and AKT. RALT suppressed AKT and ERK activities with kinetics that were significantly different from one another in distinct 32D.ErbB transfectants. However, such inhibitory function of RALT was in all cases strictly dependent on the integrity of the EBR module and, by inference, required physical interaction and spatial segregation of RALT with activated ErbB receptors.
Impact of RALT on mitogenic signals generated by ErbB RTKs
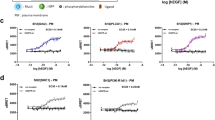
We next studied the impact of RALT and RALT ΔEBR signalling on mitogenic programs governed by ErbB RTKs. Ectopically expressed RALT did not influence the ability of 32D derivatives to proliferate in response to IL-3 (data not shown). In contrast, EGF-dependent proliferation of 32D.E1 RALT cells was severely reduced in comparison to their Pinco counterpart (Figure 9a). The suppressive effect of RALT on EGFR mitogenic signals was almost complete at suboptimal concentrations of EGF; saturating doses of EGF alleviated RALT-mediated suppression, rescuing mitogenic activity to 25–30% of that of Pinco cells (Figure 9a). Similar results were obtained with TGF-α (data not shown). Annexin V staining indicated that EGF stimulation rescued 32D.E1 Pinco cells from apoptotic cell death induced by IL-3 deprivation. In contrast, EGF-treated 32D.E1 RALT cells underwent apoptosis at rates similar to those of 32D.E1 Pinco and 32D.E1 RALT cells cultured in medium devoid of both EGF and IL-3 (data not shown). Thus, in 32D.E1 cells RALT inhibits EGFR signals necessary for both cell survival and cell proliferation, but does not exert a direct proapoptotic activity. The RALT ΔEBR mutant showed a bimodal behavior. At concentrations of EGF equal to or lower than 3 ng/ml, expression of RALT ΔEBR partially suppressed the mitogenic responses of 32D.E1 cells (Figure 9a). On the other hand, at EGF concentrations higher than 3 ng/ml, which in this assay corresponded approximately to the ED50 of EGF, the ΔEBR mutant enhanced EGF-dependent proliferation of 32D.E1 cells.
Impact of RALT signalling on mitogenic activity of defined ErbB dimers. Pinco, RALT and RALT ΔEBR derivatives of 32D.E1 (a), 32D.E2.E3 (b) and 32D.E4 (c) transfectants described in Figure 6 were seeded at 1 × 104 cells/well in IL-3-free medium and either kept in IL-3-free medium or supplemented with either EGF (a) or NRG-1β (b, c) to the indicated final concentrations; control wells were added IL-3. After 44 h, cells were pulse-labelled for 4 h with 1 μCi/ml [3H]methyl-thymidine. EGF- and NRG-1β-dependent cell proliferation was calculated as percentage over IL-3-dependent proliferation. A representative experiment of four independent assays for E1, E2.E3 and E4 transfectants is shown. Each experimental point was determined in quadruplicate wells, with standard deviation not exceeding 15%
Expression of RALT in 32D.E2.E3 cells led to the suppression of mitogenic responses elicited by NRG-1 (Figure 9b). The suppressive activity of RALT was strongest at concentrations of NRG-1 equal to or lower than its ED50, with higher NRG-1 doses somehow overcoming the inhibitory activity of RALT. RALT activity prevented the NRG-1-dependent rescue of 32D.E2.E3 cells from the apoptotic effect of IL-3 deprivation, but was not proapoptotic per se (data not shown). Strikingly, RALT ΔEBR was as efficient as RALT in suppressing NRG-1-driven proliferation of 32D.E2.E3 cells at all of the NRG-1 concentrations tested, including doses in excess of those yielding optimal proliferation of 32D.E2.E3 Pinco cells (Figure 9b). Expression of RALT in 32D.E4 cells led to complete suppression of NRG-1-dependent cell proliferation (Figure 9c), even at high ligand concentrations. Similar results were obtained with β-cellulin (data not shown). As observed with E1 and E2.E3 transfectants, RALT did not exert a direct proapoptotic activity (data not shown). The ΔEBR mutation deprived RALT of the ability to suppress proliferation of 32D.E4 cells over a wide range of NRG-1 concentrations (Figure 9c). In fact, at optimal ligand concentrations, RALT ΔEBR significantly enhanced NRG-1-dependent proliferation of 32D.E4 cells.
Discussion
Quantitative aspects of RTK outputs are important in establishing the identity of receptor signals and restraining their oncogenic potential. They are defined by the balanced interplay between mechanisms of signal generation and signal extinction. Experimental manipulations leading to either ablation (Fiorini et al., 2002) or enhancement of RALT activity (Fiorentino et al., 2000) in murine fibroblasts are consistent with RALT being physiologically involved in negative signalling to the ErbB-2 receptor. Strikingly, RALT appears to be a rather selective inhibitor, as its overexpression is unable to suppress cell proliferation driven by PDGF, bFGF (Figure 1) and serum (Fiorentino et al., 2000), or cell transformation induced by v-src and v-ras (Fiorentino et al., 2000). This contrasts with the relative promiscuity of other receptor-proximal feedback inhibitors, such as mSprouty and SOCS proteins (reviewed in Fiorini et al., 2001). The aim of this study was to analyse how RALT function impacts on the activity of the ErbB network.
Physical interaction of RALT with ErbB receptors
RALT is able to form a physical complex in living cells with ligand-activated ErbB RTKs, including EGFR, ErbB-4 and the high-affinity NRG-1 receptor generated by ErbB-2.ErbB-3 dimerization. The ErbB-binding activity of RALT can be reconstituted in vitro using a minimal 47 amino-acid region that binds distinct ErbB family members with comparable affinities.
Binding of RALT to the EGFR in intact cells requires that the receptor be catalytically active, as demonstrated by the failure of kinase-inactive EGFR to form a complex with RALT. Furthermore, the most distal 214 residues of the EGFR are dispensable for the RALT/EGFR interaction. Hence, autophosphorylation is not required for EGFR to recruit RALT and RALT must bind to the EGFR upstream of position 972.
The requirements of ErbB-1 for RALT binding recapitulate those described for ErbB-2/RALT complex formation (Fiorentino et al., 2000) and may in fact underlie the entire spectrum of ErbB/RALT interactions. Thus, the interaction of RALT with ErbB-4 is also dependent on ligand activation of the receptor, while ErbB-1 and ErbB-2 may bind RALT in a ligand-independent fashion when constitutively activated. Conversely, catalytically impaired ErbB-3 cannot bind RALT directly and is found in a complex with RALT only if recruited into a heterodimer with ErbB-2.
Recently, in structure–function studies based on the yeast two-hybrid assay Hackel et al. (2001) found that RALT is unable to bind to kinase-inactive EGFR. These authors also suggested that RALT binds to the sequence comprised between positions 985–995 of activated EGFR. This conclusion was based on the observation that EGFR baits spanning positions 646–995 were able to trans-activate reporter genes in a RALT-dependent fashion, an activity that was lost in the 646–985 and 640–940 EGFR baits (Hackel et al., 2001). In contrast, our coimmunoprecipitation experiments show that EGFR truncation at position 972 still allows efficient recruitment of RALT (Figure 5c). The latter result is compatible with yeast two-hybrid experiments that provisionally delimited the RALT-binding surface of ErbB-2 to the NH2 half of its kinase domain, that is, well apart from the ErbB-2 COOH tail (Fiorentino et al., 2000). Clearly, only the precise mapping of the RALT-binding surface in different ErbB RTKs will allow to solve these discrepancies. Based on our data, we propose that RALT binds to the catalytic domain of kinase-active ErbB RTKs following a ligand-induced conformational change of the receptor. This model is consistent with the inability of RALT to be recruited to catalytically impaired ErbB-3 in the absence of ErbB-2. Upon triggering of ErbB RTKs, RALT molecules undergo redistribution from a cytosolic location to a vesicular compartment, which likely corresponds to endocytic vesicles. This process shows an absolute requirement for the integrity of the EBR module of RALT. Spatial segregation with ligand-engaged ErbB receptors appears to be directly linked to some aspects of RALT signalling to the ErbB network, that is, suppression of activation of ERKs and AKT (see below for further discussion).
RALT is a negative regulator of mitogenic signals generated by the ErbB signalling network
In murine fibroblasts, RALT suppresses mitogenic signals generated by a recombinant EGFR.ErbB-2 chimeric receptor (Fiorentino et al., 2000). Under physiological conditions, the signalling activity of ErbB-2 is dictated by its lateral interactions with other ErbB family members. We report here that RALT suppresses the mitogenic activity of NRG-1-activated ErbB-2.ErbB-3 dimers as well as that of EGFR and ErbB-4 homodimers. RALT suppresses mitogenic signalling by ErbB-2.ErbB-4 dimers activated by either EGF or NRG-1 (our unpublished observation) as well as the transforming activity of overexpressed EGFR in Rat1 fibroblasts (Hackel et al., 2001). Thus, RALT affects signalling by all ErbB RTK combinations tested so far and it does so in several cellular model systems and different biological assays. These findings, along with the demonstration that RALT expression is induced by growth factors capable of activating different ErbB RTKs (Fiorentino et al., 2000) are consistent with RALT being a feedback inhibitor of the entire ErbB signalling network. Comparable levels of RALT suppressed the mitogenic activity of ErbB-4 much more efficiently than that of E2.E3 dimers. This was mirrored by remarkable differences in the ability of RALT to interfere with E4 and E2.E3 signals leading to ERK and AKT activation (see below for further discussion). The potency of RALT signalling to EGFR could not be directly compared to RALT activity towards E4 and E2.E3, given the lower level of RALT expression in 32D.E1 cells. We notice, however, that NRG-driven proliferation of 32D.E4 cells was ablated by RALT activity even when cells were cultured with high concentrations of NRG-1. In contrast, saturating doses of ligand partially alleviated RALT suppression of E1 and E2.E3 mitogenic signals. Thus, the sensitivity of EGFR to RALT suppressive activity resembled that of E2.E3 dimers rather than that of E4.
The differential sensitivity of ErbB RTKs to RALT signalling may impact on signal specification by the ErbB network whenever a promiscuous ligand activates more than one type of ErbB dimers in the same target cell. For instance, NRG-1-driven RALT expression may be used to tune signals generated by the ErbB-2.ErbB-3 combination, while aborting the function of concomitantly activated ErbB-4 homodimers.
Role of RALT in determining strength and duration of AKT and ERK activity
During development, distinct thresholds of RTK-induced ERK activity are instrumental in translating morphogenetic gradients into correspondingly graded transcriptional responses (reviewed in Hazzalin and Mahadevan, 2002). In proliferating cells, growth factor stimulation and ensuing RTK activity are required throughout the G1 phase of the mitotic cell cycle (reviewed in Jones and Kazlauskas, 2001). Besides regulating transcriptional responses (Cook et al., 1999; Kops et al., 2002; Murphy et al., 2002) the Ras-ERK and PI-3K-AKT pathways couple RTK activity to the regulation of the cell cycle machinery (reviewed in Lawlor and Alessi, 2001; Pruitt and Der, 2001). Thus, intensity and duration of Ras-ERK and PI-3K-AKT signals concur in defining the biological outcome of ErbB activity in normal and transformed cells (Zhou et al., 2001a, 2001b; Neve et al., 2002; Shin et al., 2002; Viglietto et al., 2002).
Reconstitution of RALT expression in 32D cells leads to suppression of EGFR, ErbB-4 and ErbB-2.ErbB-3 signals culminating in activation of ERKs and AKT. However, the RALT-enforced suppression of ERK and AKT activity displays temporal profiles that are different in distinct 32D.ErbB transfectants. RALT expression in 32D.E4 cells led to an abrupt and robust inhibition of ERK and AKT activity since the inception of receptor triggering. In contrast, suppression of ERK and AKT activation by RALT in E1 and E2.E3 cells was clearly observed only at later stages of receptor stimulation. Additionally, the timing of suppression of ERKs and AKT was further delayed in E2.E3 cells as compared to EGFR transfectants. Delayed suppression of ErbB-dependent signals by RALT is not confined to the 32D cell background: RALT overexpression caused delayed suppression of ERK activity triggered by an EGFR/ErbB-2 chimeric receptor expressed in murine fibroblasts, while leaving serum- and PMA-driven responses unaffected (Fiorentino et al., 2000).
Our results indicate that RALT activity impacts in a selective fashion on strength and duration of ERK and AKT signals generated by E1, E4 and E2.E3 dimers. While it is not yet clear how RALT discerns among different ErbB RTKs, these findings suggest that RALT may protect cells against perturbations induced by additive or superadditive activation of these pathways. As an example, the buffering function of RALT is likely to prevent further boosting of ERK and AKT activity generated by E2.E3 dimers, should ErbB-4 be concomitantly activated.
Mechanisms of suppression of ErbB RTKs signalling by RALT
Initial studies modelled RALT as an adaptor that links cytosolic effectors to activated ErbB receptors, thereby suppressing receptor-proximal events (Fiorentino et al., 2000; Hackel et al., 2001). In order to validate this model, we have compared the biochemical and biological activities of RALT to those of RALT ΔEBR, a mutant which retains the property to couple to SH3-containing effectors but is unable to complex with and be spatially regulated by ErbB RTKs. Our data indicate that the ΔEBR mutation deprives RALT of its ability to suppress ErbB-dependent activation of ERKs and AKT. We conclude that RALT must be recruited to ErbB RTKs in order to interfere with these signalling pathways.
Since RALT inhibits multiple signalling pathways once relocated to ErbB RTKs, we infer that it acts as a general suppressor of receptor-proximal events. Indeed, Hackel et al. (2001) have reported that overexpression of RALT enhances ligand-dependent downregulation of the EGFR. Several SH3-containing proteins play essential roles in receptor endocytosis, including Grb-2, intersectin, syndapin and endophilin (Simpson et al., 1999). Thus, RALT may suppress ErbB RTKs by coupling them to elements of the endocytic machinery such as Grb-2 (Fiorentino et al., 2000) and intersectin (our unpublished data). This model would provide a rationale for the seemingly paradoxical enhancement of EGFR and ErbB-4 mitogenic activity observed upon expression of RALT ΔEBR in 32D.E1 and 32D.E4 cells (Figure 9). Such a dominant-negative effect may be explained by RALT ΔEBR acting as a cytosolic trap for spatially regulated components of the endocytic machinery.
RALT ΔEBR is able to suppress the mitogenic activity of ErbB-2.ErbB-3 dimers over a wide range of ligand concentrations and under conditions that generate normal profiles of ERK and AKT signals. We propose that the ΔEBR mutant intercepts, at a receptor-distal level, signals yet unidentified but necessary to the mitogenic activity of E2.E3 dimers. It is likely that these signals are generated in parallel to the ERK and AKT pathways and eventually converge with the latter to fully implement the E2.E3 mitogenic program. Intriguingly, the EBR-independent component of RALT signalling impacts on the mitogenic activity of E1 and E4 only at low levels of receptor occupancy. We hypothesize that high levels of EGFR and ErbB-4 activity relieve the dependency for signals intercepted by RALT ΔEBR, thus allowing the above-described dominant-negative activity of RALT ΔEBR to be unmasked.
Are there structural determinants of RALT that could account for EBR-independent signals? RALT has been reported to bind to 14-3-3 proteins (Makkinje et al., 2000), which are implicated in Q2nuclear–cytoplasmic traffic. We have found that both RALT and RALT ΔEBR accumulate in the nuclei of cells treated with leptomycin B (unpublished data). Intriguingly, a RALT fragment spanning positions 1–150 behaves as a potent transcriptional activator when fused to the DNA-binding domain of GAL-4 (unpublished data). Hence, dynamic regulation of its nuclear localization may confer to RALT the ability to regulate gene activity during G1 progression in an EBR-independent fashion.
Conclusions
RALT appears to be a versatile and selective feedback inhibitor of signals propagated by ErbB RTKs. Its expression is controlled at the transcriptional and post-translational level: such an integrated control provides for timely adjustments of RALT levels and, by inference, of its signalling activity (Fiorini et al., 2002). The RALT feedback loop is therefore suited to proofread and extinguish signals generated by ErbB RTKs. The ability of RALT to integrate negative signalling to combinatorial ErbB dimers and to affect in a differential manner strength and duration of signals generated by distinct ErbB RTKs is likely to be a mechanism through which the identity of ErbB signals is preserved and global output from the ErbB network is tightly controlled.
Materials and methods
Materials
EGF was from Upstate Biotechnology, NRG-1β, bFGF and PDGF were from R&D, glutathione–agarose, IPTG, puromycin and transferrin were from Sigma. Tissue culture media and sera were from Life Technologies.
Recombinant DNA methodologies
pcDNA3 vectors for wt EGFR, EGFR KD and EGFR ΔC have been described (Levkowitz et al., 1998) and were obtained from Y. Yarden. LTR-2-based expression vectors for ErbB-1–ErbB-4 have been previously described (Di Fiore et al., 1990; Alimandi et al., 1995, 1997). cDNA fragments encoding discrete portions of RALT EBR were generated by polymerase chain reaction (PCR) amplification using synthetic oligonucleotide primers of 22 bases. Amplification products contained 5′ BamHI and 3′ EcoRI tags to allow cloning into the pGEX 4 T vector (Pharmacia) and expression in Escherichia coli. The ΔEBR mutant was generated by fusing an EcoRI–StuI fragment of RALT cDNA encompassing aa. 1–313 to a PCR-generated StuI–XbaI fragment spanning positions 362–459 of RALT. Sequence analysis confirmed the fidelity of PCR amplification and that the fusion maintained the correct ORF skipping aa. 315–361. For expression in mammalian cells, RALT ΔEBR was cloned in Pinco vector (Fiorentino et al., 2000).
Cell lines, cell culture conditions and gene transfer procedures
NIH-3T3 cells and their derivatives were grown in Dulbecco's minimal essential medium (D-MEM) containing 10% (vol/vol) newborn calf serum (NCS). 293 human embryonal kidney cells were grown in D-MEM containing 10% fetal calf serum (FCS). Transfections of 293 cells were performed as described (Fiorentino et al., 2000). Cells were processed for immunoprecipitation experiments 36–48 h post-transfection. NIH 3T3, NIH-EGFR and NIH-EGFR/ErbB-2 cells were infected with Pinco, Pinco-RALT and Pinco–RALT ΔEBR retrovirus stocks to obtain Pinco, RALT and RALT ΔEBR derivatives. Flow-cytometry analysis of GFP-positive cells indicated that infection efficiencies were routinely >95%. 32D cells and their derivatives were cultured in RPMI medium supplemented with 10% FCS and 5% (vol/vol) conditioned medium from WEHI cells as source of IL-3. Expression of ectopic RALT and RALT ΔEBR in 32D.E1 (Di Fiore et al., 1990), 32D.E4 (Tang et al., 1998) and 32D.E2.E3 cells (Alimandi et al., 1997) was achieved by electroporation of Pinco–RALT and Pinco–RALT ΔEBR vectors as described (Alimandi et al., 1997). Transfected cells were selected in 1 μg/ml puromycin. Proliferation assays with 32D cells were performed as described (Alimandi et al., 1997). Briefly, cells were seeded at 1 × 104 cells/well in 96 flat bottom well plates in IL-3-free medium containing either ErbB ligands or 5% WEHI medium. Control cells were cultivated in IL-3-free medium. After 44 h, cells were pulsed for 4 h with 1 μCi/ml [3H]methyl-thymidine and TCA-precipitated radioactivity measured in a scintillation counter. Assays were performed in quadruplicate wells. ErbB-driven cell proliferation was calculated as percentage over IL-3-driven proliferation. Mitogenic assays of NIH-EGFR/ErbB-2 cells were performed as described (Fiorentino et al., 2000). Briefly, 5 × 103 cells/well were seeded in 24-well plates and subjected to three cycles of infection with either Pinco or PincoRALT retrovirus stocks over a 24 h period. Cells were then switched to serum-free medium containing defined growth factors. Control wells were added to serum-free medium only. Cell proliferation was assessed after 48 h by pulsing cells for 4 h with 1 μCi/ml [3H]methyl-thymidine and counting TCA-precipitated radioactivity in a scintillation counter.
Immunochemical procedures
For immunoblot analysis, cells were lysed as described (Fiorentino et al., 2000). The S1 antiserum and the 19C5/4 monoclonal antibody were used at 2 μg/ml. Anti-ERK, anti-p-ERK, anti-AKT and anti-p-AKT antibodies (New England Biolabs), anti-P-Tyr MoAb 4G10 (UBI), anti-SHC antiserum (Transduction Laboratories) and affinity-purified anti-ErbB receptors antibodies (Santa Cruz Biotechnology) were used at 1 μg/ml. GST pull-down assays were performed as described (Fiorentino et al., 2000). For coimmunoprecipitation assays, lysates were incubated for 2 h at 4°C with MoAb 19C5/4 bound to ImmunoPure resin (Pierce).
Immunofluorescence and confocal microscopy analysis
Cultures of NIH-EGFR and NIH-EGFR/ErbB-2 cells infected with Pinco–RALT and Pinco–RALT ΔEBR recombinant retroviruses were deprived of serum for 24 h prior to stimulation with EGF (10 ng/ml) for different lengths of time. Cells were fixed with 4% (vol/vol) paraformaldehyde in PBS for 10 min at 20°C and permeabilized with 0.25% (vol/vol) Triton X-100 in PBS for 10 min. Comparable results were obtained when cells were fixed and permeabilized with a 1 : 1 methanol/acetone solution for 10 min at −20°C. Immunostaining was performed with MoAb 19C5/4 (for RALT proteins), MoAb 108 (for EGFR) and polyclonal antibody M6 (for the EGFR/ErbB-2 chimera) as described (Fiorentino et al., 2000). Secondary antibodies (FITC- and TRITC-conjugated donkey anti-rabbit, anti-goat and anti-mouse antibodies) were from Jackson Immunoresearch. Samples were routinely examined with a Zeiss microscope equipped with 40 × and 50 × water-immersion objectives. Confocal analysis was carried out with a Leica NBT system, equipped with 40 × 1.00–0.5 and 100 × 1.3–0.6 oil-immersion lenses.
References
Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Di Fiore PP and Kraus MH . (1995). Oncogene, 10, 1813–1821.
Alimandi M, Wang LM, Bottaro D, Lee CC, Kuo A, Frankel M, Fedi P, Tang C, Lippman M and Pierce JH . (1997). EMBO J., 16, 5608–5617.
Carraway III KL, Sliwkowski MX, Akita R, Platko JV, Guy PM, Nuijens A, Diamonti AJ, Vandlen RL, Cantley LC and Cerione RA . (1994). J. Biol. Chem., 269, 14303–14306.
Carraway III KL, Weber JL, Unger MJ, Ledesma J, Yu N, Gassmann M and Lai C . (1997). Nature, 387, 512–516.
Cook SJ, Aziz N and McMahon M . (1999). Mol. Cell. Biol., 19, 330–341.
Di Fiore PP, Segatto O, Taylor WG, Aaronson SA and Pierce JH . (1990). Science, 248, 79–83.
Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, Alema S, Benedetti P and Segatto O . (2000). Mol. Cell. Biol., 20, 7735–7750.
Fiorini M, Alimandi M, Fiorentino L, Sala G and Segatto O . (2001). FEBS Lett., 490, 132–141.
Fiorini M, Ballaro C, Sala G, Falcone G, Alema S and Segatto O . (2002). Oncogene, 21, 6530–6539.
Freeman M . (2000). Nature, 408, 313–319.
Ghiglione C, Perrimon N and Perkins LA . (1999). Dev. Biol., 205, 181–193.
Graus-Porta D, Beerli RR, Daly JM and Hynes NE . (1997). EMBO J., 16, 1647–1655.
Graus-Porta D, Beerli RR and Hynes NE . (1995). Mol. Cell. Biol., 15, 1182–1191.
Guy PM, Platko JV, Cantley LC, Cerione RA and Carraway III KL . (1994). Proc. Natl. Acad. Sci. USA, 91, 8132–8136.
Hackel PO, Gishizky M and Ullrich A . (2001). Biol. Chem., 382, 1649–1662.
Hazzalin CA and Mahadevan LC . (2002). Nat. Rev. Mol. Cell. Biol., 3, 30–40.
Jones SM and Kazlauskas A . (2001). FEBS Lett., 490, 110–116.
Karunagaran D, Tzahar E, Beerli RR, Chen X, Graus-Porta D, Ratzkin BJ, Seger R, Hynes NE and Yarden Y . (1996). EMBO J., 15, 254–264.
Kops GJ, Medema RH, Glassford J, Essers MA, Dijkers PF, Coffer PJ, Lam EW and Burgering BM . (2002). Mol. Cell. Biol., 22, 2025–2036.
Lawlor MA and Alessi DR . (2001). J. Cell Sci., 114, 2903–2910.
Lee KF, Simon H, Chen H, Bates B, Hung MC and Hauser C . (1995). Nature, 378, 394–398.
Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B and Yarden Y . (1998). Genes Dev., 12, 3663–3674.
Lonardo F, Di Marco E, King CR, Pierce JH, Segatto O, Aaronson SA and Di Fiore PP . (1990). New Biol., 2, 992–1003.
Makkinje A, Quinn DA, Chen A, Cadilla CL, Force T, Bonventre JV and Kyriakis JM . (2000). J. Biol. Chem., 275, 17838–17847.
Marshall CJ . (1995). Cell, 80, 179–185.
Murphy LO, Smith S, Chen RH, Fingar DC and Blenis J . (2002). Nat. Cell Biol., 4, 556–564.
Neve RM, Holbro T and Hynes NE . (2002). Oncogene, 21, 4567–4576.
Olayioye MA, Beuvink I, Horsch K, Daly JM and Hynes NE . (1999). J. Biol. Chem., 274, 17209–17218.
Olayioye MA, Graus-Porta D, Beerli RR, Rohrer J, Gay B and Hynes NE . (1998). Mol. Cell. Biol., 18, 5042–5051.
Olayioye MA, Neve RM, Lane HA and Hynes NE . (2000). EMBO J., 19, 3159–3167.
Perrimon N and McMahon AP . (1999). Cell, 97, 13–16.
Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M and Yarden Y . (1996). EMBO J., 15, 2452–2467.
Pruitt K and Der CJ . (2001). Cancer Lett., 171, 1–10.
Riese DJ and Stern DF . (1998). Bioessays, 20, 41–48.
Riese DJ, van Raaij TM, Plowman GD, Andrews GC and Stern DF . (1995). Mol. Cell. Biol., 15, 5770–5776.
Shin I, Yakes FM, Rojo F, Shin NY, Bakin AV, Baselga J and Arteaga CL . (2002). Nat. Med., 8, 1145–1152.
Simpson F, Hussain NK, Qualmann B, Kelly RB, Kay BK, McPherson PS and Schmid SL . (1999). Nat. Cell Biol., 1, 119–124.
Tang CK, Goldstein DJ, Payne J, Czubayko F, Alimandi M, Wang LM, Pierce JH and Lippman ME . (1998). Cancer Res., 58, 3415–3422.
Viglietto G, Motti ML, Bruni P, Melillo RM, D'Alessio A, Califano D, Vinci F, Chiappetta G, Tsichlis P, Bellacosa A, Fusco A and Santoro M . (2002). Nat. Med., 8, 1136–1144.
Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W and Ullrich A . (1995). EMBO J., 14, 4267–4275.
Waterman H and Yarden Y . (2001). FEBS Lett., 490, 142–152.
Yarden Y and Sliwkowski MX . (2001). Nat. Rev. Mol. Cell. Biol., 2, 127–137.
Zhou BP, Liao Y, Xia W, Spohn B, Lee MH and Hung MC . (2001a). Nat. Cell Biol., 3, 245–252.
Zhou BP, Liao Y, Xia W, Zou Y, Spohn B and Hung MC . (2001b). Nat. Cell Biol., 3, 973–982.
Acknowledgements
S Anastasi dedicates this work to the loving memory of his mother. We thank Y Yarden and J Sap for reagents and C Full for tissue culture work. MF is the recipient of an AIRC fellowship. This work was supported by grants from: EC (5th program) and AIRC to OS, AIRC and CNR PF- Biotecnologie to SA, AIRC to MA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Anastasi, S., Fiorentino, L., Fiorini, M. et al. Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene 22, 4221–4234 (2003). https://doi.org/10.1038/sj.onc.1206516
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.onc.1206516
Keywords
This article is cited by
-
Role of ErbB and IL-1 signaling pathways in the dermonecrotic lesion induced by Loxosceles sphingomyelinases D
Archives of Toxicology (2023)
-
Loss of MIG-6 results in endometrial progesterone resistance via ERBB2
Nature Communications (2022)
-
ERRFI1 induces apoptosis of hepatocellular carcinoma cells in response to tryptophan deficiency
Cell Death Discovery (2021)
-
Molecular insight into the affinity, specificity and cross-reactivity of systematic hepatocellular carcinoma RALT interaction profile with human receptor tyrosine kinases
Amino Acids (2021)
-
Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6
Nature Structural & Molecular Biology (2015)