Abstract
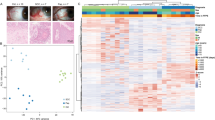
Chemical induction of squamous tumors in the mouse skin induces multiple benign papillomas: high-frequency terminally benign low-risk papillomas and low-frequency high-risk papillomas, the putative precursor lesions to squamous cell carcinoma (SCC). We have compared the gene expression profile of twenty different early low- and high-risk papillomas with normal skin and SCC. Unsupervised clustering of 514 differentially expressed genes (P<0.001) showed that 9/10 high-risk papillomas clustered with SCC, while 1/10 clustered with low-risk papillomas, and this correlated with keratin markers of tumor progression. Prediction analysis for microarrays (PAM) identified 87 genes that distinguished the two papilloma classes, and a majority of these had a similar expression pattern in both high-risk papillomas and SCC. Additional classifier algorithms generated a gene list that correctly classified unknown benign tumors as low- or high-risk concordant with promotion protocol and keratin profiling. Reduced expression of immune function genes characterized the high-risk papillomas and SCC. Immunohistochemistry confirmed reduced T-cell number in high-risk papillomas, suggesting that reduced adaptive immunity defines papillomas that progress to SCC. These results demonstrate that murine premalignant lesions can be segregated into subgroups by gene expression patterns that correlate with risk for malignant conversion, and suggest a paradigm for generating diagnostic biomarkers for human premalignant lesions with unknown individual risk for malignant conversion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Abbreviations
- DMBA:
-
dimethyl benz[a]-anthracene
- FDR:
-
false discovery rate
- K:
-
keratin
- MDS:
-
multidimensional scaling
- PAM:
-
prediction analysis for microarray
- SAM:
-
significance analysis for microarrays
- SCC:
-
squamous cell carcinomas
- TPA:
-
12-O-tetradecanoylphorbol-13-acetate
- TCR:
-
T cell receptor
- TPR:
-
tetratrico peptide repeat
- ETS:
-
E26 transformation specific
References
Balkwill F, Mantovani A . (2001). Inflammation and cancer: back to Virchow? Lancet 357: 539–545.
Brown K, Strathdee D, Bryson S, Lambie W, Balmain A . (1998). The malignant capacity of skin tumours induced by expression of a mutant H-ras transgene depends on the cell type targeted. Curr Biol 8: 516–524.
Callen JP, Bickers DR, Moy RL . (1997). Actinic keratoses. J Am Acad Dermatol 36: 650–653.
Coussens LM, Raymond WW, Bergers G, Laig-Webster M, Behrendtsen O, Werb Z et al. (1999). Inflammatory mast cells up-regulate angiogenesis during squamous epithelial carcinogenesis. Genes Dev 13: 1382–1397.
Coussens LM, Werb Z . (2002). Inflammation and cancer. Nature 420: 860–867.
Darwiche N, Scita G, Jones C, Rutberg S, Greenwald E, Tennenbaum T et al. (1996). Loss of retinoic acid receptors in mouse skin and skin tumors is associated with activation of the rasHa oncogene and high risk for premalignant progression. Cancer Res 56: 4942–4959.
de Visser KE, Korets LV, Coussens LM . (2005). De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell 7: 411–423.
Dighe AS, Richards E, Old LJ, Schreiber RD . (1994). Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity 1: 447–456.
Dong G, Loukinova E, Smith CW, Chen Z, Van Waes C . (1997). Genes differentially expressed with malignant transformation and metastatic tumor progression of murine squamous cell carcinoma. J Cell Biochem Suppl 28–29: 90–100.
Dooley TP, Reddy SP, Wilborn TW, Davis RL . (2003). Biomarkers of human cutaneous squamous cell carcinoma from tissues and cell lines identified by DNA microarrays and qRT-PCR. Biochem Biophys Res Commun 306: 1026–1036.
Dumitriu IE, Baruah P, Manfredi AA, Bianchi ME, Rovere-Querini P . (2005). HMGB1: guiding immunity from within. Trends Immunol 26: 381–387.
Girardi M, Glusac E, Filler RB, Roberts SJ, Propperova I, Lewis J et al. (2003). The distinct contributions of murine T cell receptor (TCR)gammadelta+ and TCRalphabeta+ T cells to different stages of chemically induced skin cancer. J Exp Med 198: 747–755.
Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R et al. (2001). Regulation of cutaneous malignancy by gammadelta T cells. Science 294: 605–609.
Glick AB, Kulkarni AB, Tennenbaum T, Hennings H, Flanders KC, O’Reilly M et al. (1993). Loss of expression of transforming growth factor β in skin and skin tumors is associated with hyperproliferation and a high risk for malignant conversion. Proc Natl Acad Sci USA 90: 6076–6080.
Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL . (2004). Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest 113: 913–923.
Hennings H, Shores R, Mitchell P, Spangler EF, Yuspa SH . (1985). Induction of papillomas with a high probability of conversion to malignancy. Carcinogenesis 6: 1607–1610.
Hummerich L, Muller R, Hess J, Kokocinski F, Hahn M, Furstenberger G et al. (2006). Identification of novel tumour-associated genes differentially expressed in the process of squamous cell cancer development. Oncogene 25: 111–121.
Juric D, Sale S, Hromas RA, Yu R, Wang Y, Duran GE et al. (2005). Gene expression profiling differentiates germ cell tumors from other cancers and defines subtype-specific signatures. Proc Natl Acad Sci USA 102: 17763–17768.
Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ et al. (1998). Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA 95: 7556–7561.
Larcher F, Bauluz C, Diaz-Guerra M, Quintanilla M, Conti CJ, Ballestin C et al. (1992). Aberrant expression of the simple epithelial type II keratin 8 by mouse skin carcinomas but not papillomas. Mol Carcinog 6: 112–121.
Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D et al. (2004). Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med 350: 1828–1837.
Ma C, Staudt LM . (1996). LAF-4 encodes a lymphoid nuclear protein with transactivation potential that is homologous to AF-4, the gene fused to MLL in t(4;11) leukemias. Blood 87: 734–745.
Morris RJ, Coulter K, Tryson K, Steinberg SR . (1997). Evidence that cutaneous carcinogen-initiated epithelial cells from mice are quiescent rather than actively cycling. Cancer Res 57: 3436–3443.
Morris RJ, Tryson KA, Wu KQ . (2000). Evidence that the epidermal targets of carcinogen action are found in the interfollicular epidermis of infundibulum as well as in the hair follicles. Cancer Res 60: 226–229.
Nambiar PR, Nakanishi M, Gupta R, Cheung E, Firouzi A, Ma XJ et al. (2004). Genetic signatures of high- and low-risk aberrant crypt foci in a mouse model of sporadic colon cancer. Cancer Res 64: 6394–6401.
Nishimura H, Washizu J, Nakamura N, Enomoto A, Yoshikai Y . (1998). Translational efficiency is up-regulated by alternative exon in murine IL-15 mRNA. J Immunol 160: 936–942.
Nishimura H, Yajima T, Naiki Y, Tsunobuchi H, Umemura M, Itano K et al. (2000). Differential roles of interleukin 15 mRNA isoforms generated by alternative splicing in immune responses in vivo. J Exp Med 191: 157–170.
Ozaki K, Inoue K, Sato H, Iida A, Ohnishi Y, Sekine A et al. (2004). Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-alpha secretion in vitro. Nature 429: 72–75.
Perez-Losada J, Balmain A . (2003). Stem-cell hierarchy in skin cancer. Nat Rev Cancer 3: 434–443.
Peyssonnaux C, Zinkernagel AS, Datta V, Lauth X, Johnson RS, Nizet V . (2006). TLR-4 dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood 107: 3727–3732.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6: 1–6.
Ridd K, Zhang SD, Edwards RE, Davies R, Greaves P, Wolfreys A et al. (2006). Association of gene expression with sequential proliferation, differentiation and tumor formation in murine skin. Carcinogenesis 27: 1556–1566.
Schlingemann J, Hess J, Wrobel G, Breitenbach U, Gebhardt C, Steinlein P et al. (2003). Profile of gene expression induced by the tumour promotor TPA in murine epithelial cells. Int J Cancer 104: 699–708.
Serewko MM, Popa C, Dahler AL, Smith L, Strutton GM, Coman W et al. (2002). Alterations in gene expression and activity during squamous cell carcinoma development. Cancer Res 62: 3759–3765.
Smyth MJ, Thia KY, Street SE, MacGregor D, Godfrey DI, Trapani JA . (2000). Perforin-mediated cytotoxicity is critical for surveillance of spontaneous lymphoma. J Exp Med 192: 755–760.
Tennenbaum T, Weiner AK, Belanger AJ, Glick AB, Hennings H, Yuspa SH . (1993). The suprabasal expression of α6β4 integrin is associated with a high risk for malignant progression in mouse skin carcinogenesis. Cancer Res 53: 4803–4810.
Tibshirani R, Hastie T, Narasimhan B, Chu G . (2002). Diagnosis of multiple cancer types by shrunken centroids of gene expression 2. Proc Natl Acad Sci USA 99: 6567–6572.
Yuspa SH . (1998). The pathogenesis of squamous cell cancer: lessons learned from studies of skin carcinogenesis. J Dermatol Sci 17: 1–7.
Acknowledgements
ND was supported by the American University of Beirut in Beirut/Lebanon and by the National Cancer Institute Scientist Exchange Program Fellowship. RP-L and AG were supported by the Penn State Huck Institutes of Life Sciences the Penn State Institute of the Environment, and NIH Grant CA117957 to AG This research was also supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Supplementary information
Rights and permissions
About this article
Cite this article
Darwiche, N., Ryscavage, A., Perez-Lorenzo, R. et al. Expression profile of skin papillomas with high cancer risk displays a unique genetic signature that clusters with squamous cell carcinomas and predicts risk for malignant conversion. Oncogene 26, 6885–6895 (2007). https://doi.org/10.1038/sj.onc.1210491
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.onc.1210491
Keywords
This article is cited by
-
CD271 activation prevents low to high-risk progression of cutaneous squamous cell carcinoma and improves therapy outcomes
Journal of Experimental & Clinical Cancer Research (2023)
-
Loss of epidermal MCPIP1 is associated with aggressive squamous cell carcinoma
Journal of Experimental & Clinical Cancer Research (2021)
-
Loss of DLX3 tumor suppressive function promotes progression of SCC through EGFR–ERBB2 pathway
Oncogene (2021)
-
Metabolism and genotoxicity of polycyclic aromatic hydrocarbons in human skin explants: mixture effects and modulation by sunlight
Archives of Toxicology (2020)
-
VAV2 signaling promotes regenerative proliferation in both cutaneous and head and neck squamous cell carcinoma
Nature Communications (2020)