Abstract
Penile cancer, observed only rarely in the western world, represents a carcinoma that may be cured by resection of primary lesion and in case of lymph node metastasis by early lymph node dissection. This early inguinal lymphadenectomy bares a significant better survival even in cases of nonpalpable lymph nodes, but carries also a high risk of overtreatment, especially in lower tumor stages. Due to the low incidence, only few data are available on the molecular genetic background of this tumor, especially concerning tumor progression and metastasis. Therefore, we studied 62 microsatellite markers in 28 penile carcinomas searching for markers predicting progression or outcome. LOH in more than 25% of primary tumors was found on six different chromosomes, including 2q, 6p, 8q, 9p, 12q and 17p13. Statistically significant correlations could be established in D6S260 to clinical outcome and in markers from chromosomes 6, 9 and 12 to tumor stage and metastasis. These regions are worthy for further analysis concerning tumor suppressor genes and metastasis suppressor genes.
Similar content being viewed by others
Main
Penile squamous cell carcinoma (PSCC) is an uncommon tumor entity in North America and Europe (incidence of 1/100 000). PSCC is characterized by a slow regional tumor progression and frequent metastases of inguinal lymph nodes. The incidence of lymph node metastasis varies from 0–30% in T1 tumors to 25–50% in T2–T3 stages1 and has been established as the main variable for the survival of patients. PSCC may be cured by resection of primary lesion and involved regional lymph nodes, but inguinal lymphadenectomy is associated with a high risk for complications, such as wound infection, necrosis and moderate to severe lymphedema and a higher mortality.2, 3 Therefore, accurate diagnosis of metastasis is required and the detection of reliable markers for the occurrence of metastasis would result in a great benefit for the patients. In this context, several histopathological factors of the primary penile tumor have been discussed, for example tumor stage, histopathological growth pattern, grade of differentiation, depth of invasion and presence of angioinvasion.3, 4, 5, 6, 7 Additionally, the over- or underexpression of certain proteins has been examined for its importance in tumor progression. Martins et al8 reported a prognostic significance of tumor suppressor gene p53 and a significant correlation between proliferation cell nuclear antigen and lymph node metastasis. A significant shorter survival in patients with positive p53 immunoreaction of their tumors in combination with detection of HPV DNA has also been demonstrated.9 Nevertheless, only few studies searched for DNA aberrations in penile carcinoma. Alves et al10 presented deletions in 13q21–22 and 4q21–32 by means of comperative genomic hybridization. Humbey et al11 found genetic alterations in exon 4 of the p53 region. No further information about genetic imbalances in penile carcinomas is known until now. Therefore, we studied 62 microsatellite repeats from 11 different chromosomes in 28 penile carcinomas and 10 corresponding metastases for allelic imbalances and loss of heterozygosity (LOH) to search for molecular genetic characteristics with importance for progression and clinical outcome.
Materials and methods
Tumors and Patients
Twenty-eight nonselected primary penile carcinomas comprising 19 conventional squamous cell carcinomas (12 keratinizing, 7 nonkeratinizing), and 9 variants of squamous cell carcinoma, including 4 basaloid carcinomas, 2 verrucous carcinomas and 1 condylomatous, 1 papillary, 1 sarcomatoid carcinoma, as well as 10 metastases of 9 tumors and corresponding normal tissues were analyzed in this study. All specimens underwent additional independent histopathological review (CP, GSch). Histological typing was performed according to the criteria of the WHO.12 Staging of all carcinomas was performed according to the criteria proposed by the UICC13 (six pT1N0M0, three pT2N0M0, three pT3N0M0, one pT1N3M0, two pT1N1M1, one pT1N3M1, one pT2N1M0, one pT2N3M0, one pT2N1M1, one pT2N2M1, one pT3N1M0, three pT3N2M0, two pT3N2M1, one pT3N3M1 and one pT4N2M0). There was no resection of distant metastases in patients of our study with the exception of one cutaneous metastasis, which was included in DNA analysis. The mean age of the patients was 69 years (range 40–89 years). The clinicopathological data of the patients including follow-up data if available and treatment are summarized in Table 1. Surveillance of the patients was performed quarterly by clinical examination and computer tomography of abdomen and pelvis every 6 months.
DNA Isolation
DNA isolation from paraffin-embedded tissues was performed as follows. First, hematoxylin eosin-stained slides were carefully inspected by light microscopy to identify areas that carry a sufficient amount (at least 3 mm2) of tumor measured by a scaled optical adjustment. This same area was then identified on the unstained 10 μM dewaxed, rehydrated and air-dried tissue section, which was fixed in an optical installation allowing the separate isolation of predominantly neoplastic tissue without adherent nontumorous structures under microscopical control with a cannula (used for intravenous injections) as previously described.14, 15
Molecular Genetic Analysis
The 62 PCR primer pairs amplifying informative dinucleotide repeat microsatellite loci that were investigated for allelic imbalances are as follows: D2S102 (2q36.1); D3S1076 (3p21.1), D3S2456 (3p21.3) and D3S1289 (3p14.3); D4S2639 (4p), D4S3243 (4q21.2), D4S2361 (4q21.2), D4S1625 (4q31.2), D4S1629 (4q32.1) and D4S2623 (4q25); D6S1617 and D6S344 (6p25), D6S260 and D6S1267 (6p22.3), D6S273 (6p21.3), D6S1549 (6p21.1), D6S308 (6q24.1), D6S311 (6q24.3), D6S305 (6q26); D8S166 (8q12.1), D8S251 and D8S164 (8q21.1), D8S199 (8q24.1); D9S1604, D9S1748, D9S161, D9S286, D9S162, and D9S171 (9p21, covering the p16 region); D12S64 (12q21.2), D12S101 (12q22), D12S1706 (12q23.1), D12S105 (12q24.1), D12S184 (12q24); D13S787 (13q12.1), D13S1439 (13q13.3), D13S800 (13q22.1), D13S317 (13q31.1), D13S796 (13q33.3); D15S1028 (15q15.3–21.1), D15S119 and D15S1016 (15q21.1), D15S1049 (15q21.2), D15S117 (15q21.3), D15S211 (15q22), D15S1011 (15q22.2), D15S122 (15q22.3); D17S513 (17p13), D17S786 and D17S952 (17p, around p53); D18S1144 (18q21.3), D18S1148 (18q21.3), D18S19 (18q22.1), D18S1092 (18q22.1), D18S1106 and D18S1161 (18q22.3). Primer sequences were obtained from Genome Data Base (http://www.gdb.org), cytogenetic locations are according to Ensembl (http://www.ensembl.org). PCR amplification was performed in multiplex assays with fluorochrome-labeled primers (6-FAM, JOE or TAMRA) in 12.5 ml sample volumes with 2–5 ng of genomic tumor or normal DNA as template in 15 mM Tris/HCl, 50 mM KCl, with 200 μM dNTPs, 1.5 mM MgCl2, 0.1 nM primers and 1 U HotStart Taq Polymerase (Qiagen, Hilden, Germany). An initial denaturation and activation step of 12 min at 95°C was followed by 30–35 cycles of 1 min at 95°C, 1 min at 55–58°C and 2 min at 72°C, and a 30-min final elongation step at 72°C. PCR products were analyzed on an ABI310 genetic analyzer (Applied Biosystems, Darmstadt, Germany) with ROX-labeled internal lane standard. All PCR assays were repeated at least once. LOH was scored if one allele was >90% decreased in tumor DNA when compared with the same allele in normal control DNA in both PCR assays. The frequency of LOH allelic imbalance at individual markers in informative cases was classified as low (0–25% tumors with LOH), medium (26–50%) or high (>50%).
Detection of HPV DNA
The amplification of HPV 6/11 DNA with specific primer sequences F-TACACTGCTGGACAACATGC and R-GTGCGCAGATGGGACACAC and of HPV 16 DNA with specific primer sequences F-CCCAGCTGTAATCATGCATGGAGA and R-GTGTGCCCATTAACAGGTCTTCCA was detected in a duplex PCR as described by Husnjak et al.16 For the amplification of HPV 18 DNA, we used specific primer sequences F-GAATTCACTCTATGTGCAG and R-TAGTTGTTGCCTGTAGGTG as published by Riethdorf et al.17 The products were analyzed by electrophoresis on polyacrylamide gels and detected by silver staining. To prove the presence of amplificable DNA in the extractions, all of them were amplified with primers for the human betaglobin gene.
Statistical Analysis
Cramer's Phi test (χ2) for nonparametrical data was performed with the Web Chi Square Calculator, available at http://www.georgetown.edu/faculty/ballc/webtools/web_chi.html. The P-value was estimated with the Graph Pad Quickcalcs, available at http://www.graphpad.com/quickcalcs/index.cfm. Furthermore, we used the Kaplan–Meier method to plot survival function and the log–rank test to compare survival curves. A P-value of 0.05 or less was considered as statistically significant.
Results
Microsatellite Analysis in Primary Tumors
Eight of the 62 microsatellite markers were informative in less than 14 primary tumors and are therefore considered as not informative. The frequency of LOH at the remaining 54 individual markers is shown in Figure 1, indicating the number of primary tumors with LOH and the number without LOH for each marker. Forty-three markers displayed low LOH, 10 showed medium LOH, and only 1 showed high LOH. The markers with medium or high LOH were found on six different chromosomes, including 2q, 6p, 8q, 9p, 12q and 17p13 (two markers).
Allelic loss on 2q was noted in 6 of 21 (28.6%) informative cases in marker D2S102.
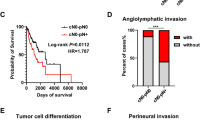
Chromosome 6 was studied with seven informative polymorphic markers, five located on 6p and two on 6q. Medium LOH was detected in 2 markers in 6p22–23, D6S260 in 7 of 22 (31.8%) informative cases and D6S1267 in 8 of 19 (42.1%) informative cases. Taken together, 12 of 28 patients (42.9%) had alterations in one or more marker from 6p22–23 in the primary tumor. Neither aberrations of all markers of chromosome 6 nor of all markers from a chromosomal arm could be found.
On the long arm of chromosome 8, four markers were investigated. D8S164 in 8q13–22.1 and D8S199 in 8q24.1 showed medium LOH (5 of 19 informative cases and 10 of 24 informative cases, respectively). The other two markers demonstrated an aberration percentage around 20%. No primary tumor, but two metastases presented losses in all four markers.
On chromosome 9, only microsatellites in the vicinity of p16/INK4A were studied. The highest frequencies of LOH were found in D9S1604 and D9S1748 (9 out of 23 (39.1%) and 10 out of 25 (40%) informative cases, respectively). Eighteen of 28 patients (64.3%) demonstrated aberrations in at least one marker.
The long arm of chromosome 12 was investigated with four informative markers. Both microsatellites from 12q13–14 showed medium LOH, whereas the other two markers located in 12q23–24 displayed an LOH frequency below 10%. In 14 of 28 patients (50%), allelic loss in 12q13–14 could be found. No case had aberrations concerning all four microsatellites from 12q. Figure 2 shows an example of allelic losses on chromosome 12.
Electropherogram of microsatellite loci from chromosome 12 in a penile primary tumor and the corresponding metastasis. The y axis represents the peak height in fluorescence units. D12S64 shows LOH in the primary tumor and in the metastasis, whereas D12S2184 is not informative. The arrows mark the lost alleles.
On chromosome 17, only the two markers around the p53 gene were informative. LOH was noted in D17S513 and D17S786 in 14 of 27 (51.9%) informative cases and 8 of 24 (33.3%) informative cases, respectively. Eighteen of 28 patients (64.3%) displayed aberrations in at least one of these two microsatellites.
Microsatellite Analysis in Metastases
Nine lymph node metastases and one cutaneous metastasis were included in this study. Seven microsatellite markers were informative in less than five metastases and were therefore considered as not informative. In general, metastatic tumors displayed more allelic imbalance than primary tumors. High LOH could be found on chromosomal arms 3p, 6p, 6q, 8q, 9p, 11q, 12q, 15q, 17p and 18q as shown in Figure 3. All allelic losses of the primary lesion were found in the corresponding metastatic lesion.
Detection of HPV DNA
In nine carcinomas, we detected HPV16 DNA (six conventional SCC, three basaloid variants). Two additional tumors showed HPV6/11 DNA (one conventional SCC, one basaloid variant), whereas HPV 18 DNA could only be demonstrated in six metastatic lesions of five different patients (four conventional SCC, one basaloid variant). Interestingly, in all basaloid variants of SCC included in this study, HPV DNA could be found.
Statistical Correlations
For the majority of markers studied, no correlation between allelic loss and clinical outcome could be established. However, LOH in D6S260 correlated significantly with shorter survival (log rank P=0.007) (Figure 4), and with regard to the markers D9S1748, D17S513 and D17S786, we observed a trend to shorter survival in patients with LOH (data not shown).
No correlation between the histological subtype of the tumor and any allelic loss could be determined with the exception of the sarcomatoid carcinoma, which displayed a high number of different aberrations compared to all other subtypes. A significant correlation between pT status and LOH was found for five different microsatellites from chromosomes 4, 6, 9, 12 and 13 (Table 2). Additionally, we detected a correlation trend with pT status for three markers from chromosomes 2, 8 and 15 (Table 2). Alterations in D2S102 and in D8S251 were observed more often in pT1, whereas LOH in the other markers was increased in pT3–4. Regarding only conventional squamous penile carcinomas, three other microsatellites from chromosomes 9, 11 and 15 correlated significantly with pT status, but only LOH in D15S119 was found more frequently in pT3–4.
The search for a relationship between allelic losses and metastasis revealed the following results. Eight microsatellites from chromosomes 6, 9, 11 and 12 correlated significantly with the occurrence of metastases (Table 2). Analysis of conventional squamous cell carcinomas only displayed a similar picture. In addition, there was a significant correlation between the detection of multiple LOH and the occurrence of metastases. Carcinomas with 10 and more affected markers showed a significant higher risk for metastasis (P=0.004).
Analysis of a possible association between HPV DNA and allelic loss showed only a significant correlation between HPV DNA-negative tumors and LOH in D9S1604 (p16INK4A region; P=0.004).
Discussion
To find prognostic parameters associated with the occurrence of metastasis, we examined genetic imbalances in penile carcinomas. The di- and tetranucleotide repeats studied were chosen due to different reasons like vicinity to known tumor-associated genes, for example, p16 or p53, the occurrence of chromosomal aberrations in a distinct region in penile carcinoma,18, 19 deletions in CGH studies of PSCC10 or frequent LOH of the marker in uterine cervix carcinoma.20, 21, 22 As there are only few molecular genetic results available on penile carcinoma so far, we applied in our study a rather broad spectrum of markers. In addition, carcinomas with similar pathomorphological characteristics like cervix carcinoma and squamous cell carcinomas of other anatomical regions demonstrate aberrations that are distributed over many chromosomal locations. To evaluate the importance of the aberrations found in our study and their impact on the carcinogenesis of PSCC, we searched for correlations between the loss of a certain marker and clinicopathological parameters like metastasis, pT status or outcome.
Allelic loss on chromosome 2q36 has already been described in cervix carcinoma21 as well as in head and neck squamous cell carcinoma.23 A variety of potential tumor suppressor genes like CASP10, BARD1, XRCC5 and PPP1R7 have been excluded as targets of mutational inactivation.21 Deletions in 2q36 have been found in early cervix carcinoma and in precancerous lesions, but in contrast to our results also in PSCC in high tumor stages. A possible explanation for this discrepancy may be the rather small number of tumors investigated here.
LOH in 6p22-23—detected in more than 30% of penile carcinoma in this study—has also been shown in cervix carcinoma,20, 24 nonsmall cell lung cancer25 or ovarian cancer,26 although no correlations to advanced stages or survival could be established. LOH in microsatellite loci D6S260 and D6S1267 is in this study frequently associated with occurrence of metastases and poor prognosis. As the corresponding 6p22–23 region is known as possible locus of newly discovered metastasis suppressor gene NOL7 in cervical carcinomas,27 this region could be useful as a prognostic marker in penile carcinomas. Further studies to determine the impact of NOL7 in penile carcinomas are in preparation.
Losses on the long arm of chromosome 8 could also be found in higher percentages in this study. LOH at 8q has been reported in oral premalignant lesions and oral cancer,28 in high percentages in squamous cell larynx cancer29 and in a variety of other tumors like pleomorphic adenoma,15 nonsmall cell lung carcinoma,30 gastric cancer31 or prostate cancer.32 Alterations of 8q have so far in most carcinomas been associated to early stages, and in our tumor samples, it was also found more often in pT1 or pT2 stages. This may indicate the impact of potential tumor suppressor genes on 8q in the onset of tumorigenesis.
Carcinomas with LOH in the region of p16INK4a (localized in the 9p21 region), frequently seen in this study, showed a significant higher risk for lymph node metastases (P=0.005). Previous examinations of vulvar carcinomas and gastric carcinomas were able to show a significant association of p16 downregulation and development of lymph node metastases.33, 34 Furthermore, the authors demonstrated a complete loss of p16 expression in case of LOH in the p16 region. As p16 is known to be involved in cell cycle regulation by inhibition of cyclin-dependent kinase 4 our data indicate an essential role of p16 alteration for development of metastases in penile carcinomas. The key role of the p16INK4a region is underlined by examinations concerning PSCC with HPV DNA presence and strong expression of p16.35 In accordance with previous findings, none of the patients of our study with LOH in D9S1604 showed HPV DNA presence.
Genetic alterations in the 18q region are frequently described for other carcinomas, for example, cervical carcinomas.20, 24 Candidate tumor suppressor gene SMAD4/DPC4 was localized in the 18q21.2 region and LOH in this region correlated with poor prognosis.36 Deleted in colon cancer (DCC) gene is another putative tumor suppressor gene located in this region. Alterations of DCC had been described for several other carcinomas than colon carcinomas, for example, renal cell carcinoma37 or nonsmall cell lung carcinoma.38 Furthermore, LOH has been associated with higher malignant stages of gastric carcinoma.39 Additionally, DCC inactivation by promoter hypermethylation has been shown in primary head and neck squamous cell carcinomas.40
Furthermore, we examined correlations of LOH in regions coding for different parts of pathways in cell cycle and carcinogenesis. Interestingly, none of the examined carcinomas showed a combination of LOH in D17S786 and D12S101 (p53 and mdm2 region, both part of the p14ARF/MDM2/p53 pathway).42, 43, 44
In addition, we were able to show a significant association between high frequency of LOH and metastasis for penile carcinomas. Carcinomas with 10 and more affected markers showed a significant higher risk for metastasis (P=0.004). This result is in accordance with the established model that metastasis is a multifactorial process.
We detected no significant differences in the molecular genetic alterations between slow growing subtypes like verrucous or papillary carcinomas and more aggressive variants like basaloid carcinoma. Interestingly, the basaloid variants showed a relative small number of LOH compared with the also poorly differentiated sarcomatoid carcinoma. A possible explanation for this result could be that sarcomatoid carcinomas resemble the more aggressive sarcomas, whereas basaloid variants still show the more structured histology of squamous carcinomas. As some authors speculate about a molecular genetic background for the differences between subtypes of squamous cell carcinomas and some molecular genetic differences have already been demonstrated,41, 42, 43, 44 further analysis of additional markers and chromosomal regions should be carried out to detect DNA aberrations that characterize aggressive variants.
In conclusion, despite the rather low number of penile carcinomas investigated, our results revealed some promising chromosomal regions that are worthy for further analysis in the search of tumor suppressor genes with impact in penile cancer.
References
Doehn C, Baumgartel M, Jocham D . Surgical therapy of penis carcinoma. Urologe 2001;A40:303–307.
Horenblas S . Lymphadenectomy for squamous cell carcinoma of the penis. Part 2: the role and technique of lymph node dissection. BJU Int 2001;88:473–483.
Ficarra V, Martignoni G, Maffei N, et al. Predictive pathological factors of lymph nodes involvement in the squamous cell carcinoma of the penis. Int Urol Nephrol 2002;34:245–250.
Solsona E, Iborra I, Rubio J, et al. Prospective validation of the association of local tumor stage and grade as a predictive factor for occult lymph node micrometastasis in patients with penile carcinoma and clinically negative inguinal lymph nodes. J Urol 2001;165:1506–1509.
Lopes A, Hidalgo GS, Kowalski LP, et al. Prognostic factors in carcinoma of the penis: multivariate analysis of 145 patients treated with amputation and lymphadenectomy. J Urol 1996;156:1637–1642.
Emerson RE, Ulbright TM, Eble JN, et al. Predicting cancer progression in patients with penile squamous cell carcinoma: the importance of depth of invasion and vascular invasion. Mod Pathol 2001;14:963–968.
Villavicencio H, Rubio-Briones J, Regalado R, et al. Grade, local stage and growth pattern as prognostic factors in carcinoma of the penis. Eur Urol 1997;32:442–447.
Martins AC, Faria SM, Cologna AJ, et al. Immunoexpression of p53 protein and proliferating cell nuclear antigen in penile carcinoma. J Urol 2002;167:89–92.
Lopes A, Bezerra AL, Pinto CA, et al. p53 as a new prognostic factor for lymph node metastasis in penile carcinoma: analysis of 82 patients treated with amputation and bilateral lymphadenectomy. J Urol 2002;168:81–86.
Alves G, Heller A, Fiedler W, et al. Genetic imbalances in 26 cases of penile squamous cell carcinoma. Genes Chromosom Cancer 2001;31:48–53.
Humbey O, Cairey-Remonnay S, Guerrini JS, et al. Detection of the human papillomavirus and analysis of the TP53 polymorphism of exon 4 at codon 72 in penile squamous cell carcinomas. Eur J Cancer 2003;39:684–690.
Eble JN, Sauter G, Epstein JI, et al. Pathology and genetics of tumours of the urinary system and male genital organs. World Health Organization Classification of Tumours. IARC Press: Lyon, 2004, pp 280–290.
Sobin LH, Wittekind C . TNM Classification of Malignant Tumours, 6th edn. Wiley-Liss: New York, 2002.
Poetsch M, Dittberner T, Woenckhaus C . PTEN/MMAC1 in malignant melanoma and its importance for tumour progression. Cancer Genet Cytogenet 2001;125:21–26.
Poetsch M, Zimmermann A, Wolf E, et al. Loss of heterozygosity occurs predominantly, but not exclusively in the epithelial compartment of pleomorphic adenoma. Neoplasia 2005;7:688–695.
Husnjak K, Grce M, Magdic L, et al. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J Virol Methods 2000;88:125–134.
Riethdorf S, Riethdorf L, Milde-Langosch K, et al. Differences in HPV 16- and HPV 18 E6/E7 oncogene expression between in situ and invasive adenocarcinomas of the cervix uteri. Virchows Arch 2000;437:491–500.
Ornellas AA, Ornellas MH, Simoes F, et al. Cytogenetic analysis of an invasive, poorly differentiated squamous cell carcinomas of the penis. Cancer Genet Cytogenet 1998;101:78–80.
Ornellas AA, Ornellas MH, Otero L, et al. Karyotypic findings in two cases of moderately differentiated squamous cell carcinomas of the penis. Cancer Genet Cytogenet 1999;115:77–79.
Kersemaekers A-MF, Kenter GG, Hermans J, et al. Allelic loss and prognosis in carcinoma of the uterine cervix. Int J Cancer 1998;79:411–417.
Narayan G, Pulido HA, Koul S, et al. Genetic analysis identifies putative tumor suppressor sites at 2q35–q36.1 and 2q36.3–q37.1 involved in cervical cancer progression. Oncogene 2003;22:3489–3499.
Elhamidi A, Hamoudi RA, Kocjan G, et al. Cervical intraepithelial neoplasia: prognosis by combined LOH analysis of multiple loci. Gynecol Oncol 2004;94:671–679.
Yamamoto N, Mizoe J, Numasawa H, et al. Allelic loss on chromosomes 2q, 3p and 21q: possibly a poor prognostic factor in oral squamous cell carcinoma. Oral Oncol 2003;39:796–805.
Kersemaekers AM, van de Vijver MJ, Kenter GG, et al. Genetic alterations during the progression of squamous cell carcinomas of the uterine cervix. Genes Chromosom Cancer 1999;26:346–354.
Tseng RC, Chang JW, Hsien FJ, et al. Genomewide loss of heterozygosity and its clinical associations in non small cell lung cancer. Int J Cancer 2005;117:214–217.
Foulkes WD, Ragoussis J, Stamp GW, et al. Frequent loss of heterozygosity on chromosome 6 in human ovarian carcinoma. Br J Cancer 1993;67:551–559.
Hasina R, Pontier AL, Fekete MJ, et al. NOL7 is a nucleolar candidate tumor suppressor gene in cervical cancer that modulates the angiogenic phenotype. Oncogene 2006;26:588–598.
Garnis C, Coe BP, Ishkanian A, et al. Novel regions of amplification on 8q distinct from the MYC locus and frequently altered in oral dysplasia and cancer. Genes Chromosom Cancer 2004;39:93–98.
Sasiadek M, Stembalska-Kozlowska A, Smigiel R, et al. Microsatellite and chromosome instability in squamous cell laryngeal carcinoma. Int J Oncol 2001;19:401–405.
Sasatomi E, Finkelstein SD, Woods JD, et al. Comparison of accumulated allele loss between primary tumor and lymph node metastasis in stage II non-small cell lung carcinoma: implications for the timing of lymph node metastasis and prognostic value. Cancer Res 2002;62:2681–2689.
Kakinuma N, Kohu K, Sato M, et al. Candidate regions of tumor suppressor gene by loss of heterozygosity analysis on chromosome 8p11.1-q13.3 in gastric cancer. Cancer Lett 2004;213:111–116.
Perinchery G, Bukuroy N, Nakajima K, et al. Loss of two new loci on chromosome 8 (8p23 and 8q12–13) in human prostate cancer. Int J Oncol 1999;14:495–500.
Knopp S, Bjorge T, Nesland JM, et al. p16INK4a and p21Waf1/Cip1 expression correlates with clinical outcome in vulvar carcinomas. Gynecol Oncol 2004;95:37–45.
He XS, Rong YH, Su Q, et al. Expression of p16 gene and Rb protein in gastric carcinoma and their clinicopathological significance. World J Gastroenterol 2005;11:2218–2223.
Ferreux E, Lont AP, Horenblas S, et al. Evidence for at least three alternative mechanisms targeting the p16INK4A/cyclin D/Rb pathway in penile carcinoma, one of which is mediated by high-risk human papillomavirus. J Pathol 2003;201:109–118.
Isaksson-Mettavainio M, Palmqvist R, Forssell J, et al. SMAD4/DPC4 expression and prognosis in human colorectal cancer. Anticancer Res 2006;26:507–510.
Hirata H, Matsuyama H, Matsumoto H, et al. Deletion mapping of 18q in conventional renal cell carcinoma. Cancer Genet Cytogenet 2005;163:101–105.
Baksh FK, Dacic S, Finkelstein SD, et al. Widespread molecular alterations present in stage I non-small cell lung carcinoma fail to predict tumor recurrence. Mod Pathol 2003;16:28–34.
Wang D, Fang D, Luo Y, et al. Study of loss of heterozygosity at DCC and APC/MCC genetic loci of gastric cancer. Chin Med Sci J 1999;14:107–111.
Carvalho AL, Chuang A, Jinag WW, et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res 2006;66:9401–9407.
Kleist B, Bankau A, Lorenz G, et al. Different risk factors in basaloid and common squamous head and neck cancer. Laryngoscope 2004;114:1063–1068.
Choi HR, Roberts DB, Johnigan RH, et al. Molecular and clinicopathologic comparisons of head and neck squamous carcinoma variants: common and distinctive features of biological significance. Am J Surg Pathol 2004;28:1299–1310.
Tonon G, Brennan C, Protopopov A, et al. Common and contrasting genomic profiles among the major human lung cancer subtypes. Cold Spring Harb Symp Quant Biol 2005;70:11–24.
Greenawalt DM, Duong C, Smyth GK, et al. Gene expression profiling of esophageal cancer: comparative analysis of Barrett's esophagus, adenocarcinoma, and squamous cell carcinoma. Int J Cancer 2007;120:1914–1921.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Poetsch, M., Schuart, BJ., Schwesinger, G. et al. Screening of microsatellite markers in penile cancer reveals differences between metastatic and nonmetastatic carcinomas. Mod Pathol 20, 1069–1077 (2007). https://doi.org/10.1038/modpathol.3800931
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/modpathol.3800931
Keywords
This article is cited by
-
Diagnostik und Therapie des Peniskarzinoms
Uro-News (2022)
-
Prognostic implications of genotyping and p16 immunostaining in HPV-positive tumors of the uterine cervix
Modern Pathology (2020)
-
Understanding genomics and the immune environment of penile cancer to improve therapy
Nature Reviews Urology (2020)
-
Peniskarzinom
Der Urologe (2020)
-
Molekulares Tumorboard Peniskarzinom – eine Herausforderung
Der Urologe (2019)