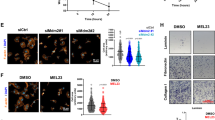
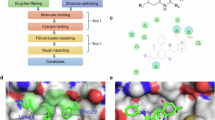
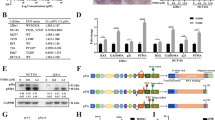
Abstract
MDM2, a ubiquitin E3-ligase of the RING family, has a key role in regulating p53 abundance. During normal non-stress conditions p53 is targeted for degradation by MDM2. MDM2 can also target itself and MDMX for degradation. MDMX is closely related to MDM2 but the RING domain of MDMX does not possess intrinsic E3-ligase activity. Instead, MDMX regulates p53 abundance by modulating the levels and activity of MDM2. Dimerization, mediated by the conserved C-terminal RING domains of both MDM2 and MDMX, is critical to this activity. Here we report the crystal structure of the MDM2/MDMX RING domain heterodimer and map residues required for functional interaction with the E2 (UbcH5b). In both MDM2 and MDMX residues C-terminal to the RING domain have a key role in dimer formation. In addition we show that these residues are part of an extended surface that is essential for ubiquitylation in trans. This study provides a molecular basis for understanding how heterodimer formation leads to stabilization of MDM2, yet degradation of p53, and suggests novel targets for therapeutic intervention.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- GST:
-
glutathione-S-transferase
- MDM:
-
mouse double minute
- RING:
-
really interesting new gene 1
References
Vousden KH, Lane DP . p53 in health and disease. Nat Rev Mol Cell Biol 2007; 8: 275–283.
Yang Y, Li CC, Weissman AM . Regulating the p53 system through ubiquitination. Oncogene 2004; 23: 2096–2106.
Shmueli A, Oren M . Regulation of p53 by MDM2: fate is in the numbers. Mol Cell 2004; 13: 4–5.
Brooks CL, Gu W . p53 ubiquitination: MDM2 and beyond. Mol Cell 2006; 21: 307–315.
Stommel JM, Wahl GM . Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 2004; 23: 1547–1556.
de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG . Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem 2003; 278: 38315–38324.
LeBron C, Chen L, Gilkes DM, Chen J . Regulation of Mdmx nuclear import and degradation by Chk2 and 14-3-3. EMBO J 2006; 25: 1196–1206.
Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A et al. DNA damage-induced phosphorylation of MDMX at serine 367 activates p53 by targeting MDMX for MDM2-dependent degradation. Mol Cell Biol 2005; 25: 9608–9620.
Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 2002; 277: 19251–19254.
Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M . HDMX stimulates HDM2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA 2003; 100: 12009–12014.
Sharp DA, Kratowicz SA, Sank MJ, George DL . Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 1999; 274: 38189–38196.
Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M . MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 1999; 447: 5–9.
Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE . Solution structure of the HDM2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol 2006; 363: 433–450.
Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM . RING domain-mediated interaction is a requirement for MDM2's E3 ligase activity. Cancer Res 2007; 67: 6026–6030.
Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 2001; 276: 14537–14540.
Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK . Structure and E3-ligase activity of the RING-RING complex of polycomb proteins Bmi1 and RING1b. EMBO J 2006; 25: 2465–2474.
Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM . MDM2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 2000; 275: 8945–8951.
Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N et al. The MDM2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 2007; 26: 90–101.
Uldrijan S, Pannekoek WJ, Vousden KH . An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J 2007; 26: 102–112.
Vander Kooi CW, Ohi MD, Rosenberg JA, Oldham ML, Newcomer ME, Gould KL et al. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry 2006; 45: 121–130.
Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C et al. Chaperoned ubiquitylation – crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell 2005; 20: 525–538.
Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE . Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol 2001; 8: 833–837.
Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM . Structure of a Bmi-1-RING1b polycomb group ubiquitin ligase complex. J Biol Chem 2006; 281: 20643–20649.
Zheng N, Wang P, Jeffrey PD, Pavletich NP . Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 2000; 102: 533–539.
Ozkan E, Yu H, Deisenhofer J . Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA 2005; 102: 18890–18895.
Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R et al. Amplification of MDMX (or MDM4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 2004; 24: 5835–5843.
Singh RK, Iyappan S, Scheffner M . Hetero-oligomerization with MDMX rescues the ubiquitin/NEDD8 ligase activity of RING finger mutants of MDM2. J Biol Chem 2007; 282: 10901–10907.
Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M et al. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci USA 2005; 102: 16182–16187.
Wang X, Taplick J, Geva N, Oren M . Inhibition of p53 degradation by MDM2 acetylation. FEBS Lett 2004; 561: 195–201.
Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindström MS et al. Targeted Inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 2007; 12: 355–366.
Stad R, Little NA, Xirodimas DP, Frenk R, van der Eb AJ, Lane DP et al. MDMX stabilizes p53 and MDM2 via two distinct mechanisms. EMBO Rep 2001; 2: 1029–1034.
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–848.
Marine JC, Dyer MA, Jochemsen AG . MDMX: from bench to bedside. J Cell Sci 2007; 120: 371–378.
Bell S, Klein C, Muller L, Hansen S, Buchner J . p53 contains large unstructured regions in its native state. J Mol Biol 2002; 322: 917–927.
Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT . Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol 2000; 20: 8458–8467.
Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002; 416: 703–709.
Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV et al. Small molecule inhibitors of Hdm2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 2005; 7: 547–559.
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; 50: 760–763.
Vonrhein C, Blanc E, Roversi P, Bricogne G . Automated structure solution with autosharp. Methods Mol Biol 2006; 364: 215–230.
Perrakis A, Morris R, Lamzin VS . Automated protein model building combined with iterative structure refinement. Nat Struct Biol 1999; 6: 458–463.
Emsley P, Cowtan K . Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60: 2126–2132.
Acknowledgements
We thank Rayleen Fredericks-Short and Nelly Olova for excellent technical assistance, Torsten Kleffmann and the Centre for Protein Research (University of Otago) for mass spectrometry analysis, Mark Hinds for helpful discussions, and Andrew Mercer and Anthony Braithwaite for reagents. The SSRL Structural Molecular Biology program acknowledges the NCRR (Grant No P41 RR001209), a component of the NIH for funding. This work was supported by the Marsden Fund (NZ) (CLD), and PDM is a recipient of a Health Sciences Career Development Award (University of Otago).
Author information
Authors and Affiliations
Corresponding author
Additional information
Data deposition: The atomic co-ordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID 2vje and PDB ID 2vjf).
Edited by: B Zhivotovsky
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Linke, K., Mace, P., Smith, C. et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 15, 841–848 (2008). https://doi.org/10.1038/sj.cdd.4402309
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.cdd.4402309
Keywords
This article is cited by
-
1H, 15N and 13C backbone resonance assignments of the acidic domain of the human MDM2 protein
Biomolecular NMR Assignments (2023)
-
Structure of the nutrient-sensing hub GATOR2
Nature (2022)
-
1H, 15N and 13C backbone resonance assignments of the acidic domain of the human MDMX protein
Biomolecular NMR Assignments (2022)
-
MDMX is essential for the regulation of p53 protein levels in the absence of a functional MDM2 C-terminal tail
BMC Molecular and Cell Biology (2021)
-
Critical role of cysteine-266 of SIE3 in regulating the ubiquitination and degradation of SIP1 transcription factor in Lotus japonicus
Planta (2021)