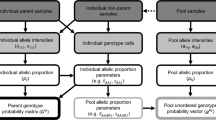
Abstract
The cost of large-scale association studies may be reduced substantially by analysis of pooled DNA from multiple individuals. Here we examine the optimal symmetric and asymmetric designs for pooling experiments for quantitative traits under a range of assumptions about the underlying genetic model and the sources of experimental errors in allele frequency estimation. The results indicate that, in the absence of experimental errors and for common alleles with additive effects, a symmetric pooling scheme comparing the top 27% with the bottom 27% of the trait distribution is optimal, extracting 80% the total information available. A symmetric design is not optimal for rare or recessive alleles, which require asymmetric (or other) pooling strategies. Allele frequency measurement errors reduce the optimal pooling fraction as well as the overall efficiency of the pooling design. In contrast, random variation in the amount of DNA contributed by individuals to a pool reduces only the overall efficiency of the pooling design. Our results emphasize the importance of minimising experimental errors and suggest a pooling fraction of around 20%.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Risch N, Merikangas K . The future of genetic studies of complex human diseases Science 1996 273: 1516–1517
Abecasis GR, Noguchi E, Heinzmann A et al. Extent and distribution of linkage disequilibrium in three genomic regions Am J Hum Genet 2001 68: 191–197
Collins FS, Guyer MS, Chakarvarti A . Variations on a theme: cataloging human DNA sequence variation Science 1997 274: 1580–1581
Barcellos LF, Klitz W, Field LL et al. Association mapping of disease loci, by use of a pooled DNA genomic screen Am J Hum Genet 1997 61: 734–747
Daniels J, Holmans P, Williams N et al. A simple method for analysing microsatellite allele image patterns generated from DNA pools and its applications to allelic association studies Am J Hum Genet 1998 62: 1189–1197
Fisher PJ, Turic D, Williams NM et al. DNA pooling identifies QTLs on chromosome 4 for general cognitive ability in children Hum Mol Genet 1999 8: 915–922
Hill L, Craig IW, Asherson P et al. DNA pooling and dense marker maps: a systematic search for genes for cognitive ability Neuroreport 1999 10: 843–848
Shaw SH, Carrasquillo MM, Kashuk C, Puffenberger EG, Chakravarti A . Allele frequency distributions in pooled DNA samples: applications to mapping complex disease genes Genome Res 1998 8: 111–123
Stockton DW, Lewis RA, Abboud EB et al. A novel locus for Leber congenital amaurosis on chromosome 14q24 Hum Genet 1998 103: 328–333
Suzuki K, Bustos T, Spritz RA . Linkage disequilibrium mapping of the gene for Margarita Island ectodermal dysplasia (ED4) to 11q23 Am J Hum Genet 1998 63: 1102–1107
Risch N, Teng J . The relative power of family-based and case-control designs for linkage disequilibrium studies of complex human diseases I. DNA pooling Genome Res 1998 8: 1273
Bader JS, Bansal A, Sham P . Efficient SNP-based tests of association for quantitative phonotypes using pooled DNA Genescreen 2002 in press
Hoogendoorn B, Norton N, Kirov G et al. Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools Hum Genet 2000 107: 488–493
Breen G, Sham PC, Li T, Shaw D, Collier D, Clair ST . Accuracy and sensitivity of DNA pooling with microsatellite repeats using capillary electrophoresis Mol Cell Probes 1999 13: 1–7
Schork NJ, Nath SK, Fallin D, Chakarvati A . Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control Subjects Am J Hum Genet 2000 67: 1208–1218
Sham, PC, Cherny SS, Purcell S, Hewitt JK . Power of linkage versus association analyses of quantitative traits, by use of variance-components models, for sibship data Am J Hum Genet 2000 66: 1616–1630
Satten GA, Flanders DW, Yang Q . Accounting for unmeasured population substructure in case-control studies of genetic association using a novel latent-class model Am J Hum Genet 2001 68: 466–477
Devlin B, Roeder K . Genomic control for association studies Biometrics 2001 55: 788–808
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P . Inference of population structure using multilocus genotype data Genetics 2000 155: 945–959
Pritchard JK, Rosenberg NA . Use of unlinked genetic markers to detect population stratification in association studies Am J Hum Genet 1999 65: 220–228
Mood AM, Graybill FA, Boes DC . Introduction to the theory of statistics McGraw-Hill Book Company 3rd edn 1974 pp. 181
Falconer DS . The inheritance of liability to certain diseases estimated from the incidence among relatives An Hum Genet 1965 51: 227–233
Acknowledgements
We would like to thank Jing Hua Zhao for helpful comments. This research was supported in part by a UK MRC research studentship to A Jawaid, UK MRC component grant G9700821, Wellcome Trust grant 055379, and National Institutes of Health grant EY-12562.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A
Variance due to unequal contribution of DNA samples
Restricting the terminology to this appendix, let Xi represent the total number of alleles contributed by individual i in a pool made up of n individuals, with Xi ∼ N(μ,τ2μ2). Let Yi represent the number of A1 alleles contributed by individual i. For genotype A1A1 with population frequency p2, Yi=Xi; for A1A2 with frequency p1, Yi ∼Bin(Xi,1/2); and for A2A2 with frequency p0, Yi=0. The population frequency of allele A1 is p=p1/2+p2, and the frequency of allele A1 in the pool is

being the approximate variance for a quotient of correlated random variables.21 The required terms are

after simplification. Assuming Hardy-Weinberg equilibrium and large μ, this reduces to

Appendix B
Optimal symmetric design in thepresence of experimental error
Let G be the proportion of A1 alleles in a genotype, so that G = 0, 1/2 or 1, and Var(G) = pq/2. According to an additive genetic model, the expected value of the trait X given G is  . Using the implied covariance, Cov(X,G)=pqa, and a linear approximation, the expected value of G given X
. Using the implied covariance, Cov(X,G)=pqa, and a linear approximation, the expected value of G given X  . In the lower pool, E(X)≈−Φ(Φ−1(f))/f, where Φ is the standard normal density function, Φ−1 is the inverse standard normal distribution function, and f is the lower pooling fraction.22 The expected values of G is in the lower pool and, by symmetry, the upper pool are therefore
. In the lower pool, E(X)≈−Φ(Φ−1(f))/f, where Φ is the standard normal density function, Φ−1 is the inverse standard normal distribution function, and f is the lower pooling fraction.22 The expected values of G is in the lower pool and, by symmetry, the upper pool are therefore

from before. The NCP is therefore

Appendix C
Analytical approximation for theoptimal symmetric design in the presence ofexperimental error
The design is optimised by maximising the value of the NCP, which is equivalent to maximising the value of y2/(f+f2κ2), where y = Φ(z) and f = Φ(z) for normal deviate z. Taking the derivative with respect to z and multiplying by non-zero terms yields

as the equation specifying the minimum. When κ = 0, the solution to this equation occurs at z0 = −0.61, with f0 = 0.27 and y0 = 0.33 (Bader et al.12). For small κ, we write z = z0 + δ. To lowest order in δ, the above equation is

When κ is large, we use the asymptotic expansion f = −(y/z) + (y/z3), and the equation specifying the optimum reduces to −2yκ2/z3 = 1. Taking the natural logarithm of both sides and equating exponents,

Writing x = − + δ yields δ = (1/8)−(1/4)ln[(2/π)1/2κ2] to lowest order in δ. The result of this perturbation theory expansion for large κ is

An appropriate crossover between the small-κ formula and the large-κ formula is κ = 1.
Rights and permissions
About this article
Cite this article
Jawaid, A., Bader, J., Purcell, S. et al. Optimal selection strategies for QTL mapping using pooled DNA samples. Eur J Hum Genet 10, 125–132 (2002). https://doi.org/10.1038/sj.ejhg.5200771
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5200771
Keywords
This article is cited by
-
A genome-wide association study of essential hypertension in an Australian population using a DNA pooling approach
Molecular Genetics and Genomics (2017)
-
Machine learning approach for pooled DNA sample calibration
BMC Bioinformatics (2015)
-
Pooling/bootstrap-based GWAS (pbGWAS) identifies new loci modifying the age of onset in PSEN1 p.Glu280Ala Alzheimer's disease
Molecular Psychiatry (2013)
-
High-resolution genetic mapping with pooled sequencing
BMC Bioinformatics (2012)
-
An optimal DNA pooling strategy for progressive fine mapping
Genetica (2009)