Abstract
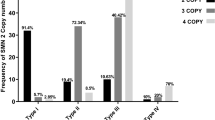
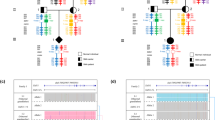
Most spinal muscular atrophy patients lack both copies of SMN1. Loss of SMN1 (‘0-copy alleles’) can occur by gene deletion or SMN1-to-SMN2 gene conversion. Despite worldwide efforts to map the segmental duplications within the SMN region, most assemblies do not correctly delineate these genes. A near pericentromeric location provides impetus for the strong evidence that SMN1 and SMN2 arose from a primate-specific paralogous gene duplication. Here we meta-analyzed our recent laboratory results together with available published data, in order to calculate new mutation rates and allele/haplotype frequencies in this recalcitrant and highly unstable region of the human genome. Based on our tested assumption of compliance with Hardy–Weinberg equilibrium, we conclude that the SMN1 allele frequencies are: ‘0-copy disease alleles,’ 0.013; ‘1-copy normal alleles,’ 0.95; ‘2-copy normal alleles (ie, two copies of SMN1 on one chromosome),’ 0.038; and ‘1D disease alleles (SMN1 with a small intragenic mutation),’ 0.00024. The SMN1 haplotype [‘(SMN1 copy number)-(SMN2 copy number)’] frequencies are: ‘0-0,’ 0.00048; ‘0-1,’ 0.0086; ‘0-2,’ 0.0042; ‘1-0,’ 0.27; ‘1-1,’ 0.66; ‘1-2,’ 0.015; ‘2-0,’ 0.027; and ‘2-1,’ 0.012. Paternal and maternal de novo mutation rates are 2.1 × 10−4 and 4.2 × 10−5, respectively. Our data provide the basis for the most accurate genetic risk calculations, as well as new insights on the evolution of the SMN region, with evidence that nucleotide position 840 (where a transition 840C>T functionally distinguishes SMN2 from SMN1) constitutes a mutation hotspot. Our data also suggest selection of the 1-1 haplotype and the presence of rare chromosomes with three copies of SMN1.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ogino S, Leonard DG, Rennert H, Ewens WJ, Wilson RB : Genetic risk assessment in carrier testing for spinal muscular atrophy. Am J Med Genet 2002; 110: 301–307.
Lefebvre S, Burglen L, Reboullet S et al: Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–165.
Burglen L, Lefebvre S, Clermont O et al: Structure and organization of the human survival motor neurone (SMN) gene. Genomics 1996; 32: 479–482.
Monani UR, Lorson CL, Parsons DW et al: A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet 1999; 8: 1177–1183.
Lorson CL, Hahnen E, Androphy EJ, Wirth B : A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA 1999; 96: 6307–6311.
Kashima T, Manley JL : A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 2003; 34: 460–463.
Wirth B : An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat 2000; 15: 228–237.
Ogino S, Wilson RB : Spinal muscular atrophy: molecular genetics and diagnostics. Expert Rev Mol Diagn 2004; 4: 15–29.
McAndrew PE, Parsons DW, Simard LR et al: Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet 1997; 60: 1411–1422.
Chen KL, Wang YL, Rennert H et al: Duplications and de novo deletions of the SMNt gene demonstrated by fluorescence-based carrier testing for spinal muscular atrophy. Am J Med Genet 1999; 85: 463–469.
Ogino S, Gao S, Leonard DG, Paessler M, Wilson RB : Inverse correlation between SMN1 and SMN2 copy numbers: evidence for gene conversion from SMN2 to SMN1. Eur J Hum Genet 2003; 11: 275–277, (see Addendum in Vol 11; 723).
Mailman MD, Hemingway T, Darsey RL et al: Hybrids monosomal for human chromosome 5 reveal the presence of a spinal muscular atrophy (SMA) carrier with two SMN1 copies on one chromosome. Hum Genet 2001; 108: 109–115.
Wirth B, Schmidt T, Hahnen E et al: De novo rearrangements found in 2% of index patients with spinal muscular atrophy: mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am J Hum Genet 1997; 61: 1102–1111.
Eichler EE, Sankoff D : Structural dynamics of eukaryotic chromosome evolution. Science 2003; 301: 793–797.
Cheung J, Estivill X, Khaja R et al: Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol 2003; 4: R25.
Ogino S, Wilson RB : Genetic testing and risk assessment for spinal muscular atrophy (SMA). Hum Genet 2002; 111: 477–500.
Ogino S, Wilson RB : SMN dosage analysis and risk assessment for spinal muscular atrophy. Am J Hum Genet 2002; 70: 1596–1598; discussion 1598–1599.
Ogino S, Leonard DG, Rennert H, Wilson RB : Spinal muscular atrophy genetic testing experience at an academic medical center. J Mol Diagn 2002; 4: 53–58.
Ogino S, Wilson RB : Quantification of PCR bias caused by a single nucleotide polymorphism in SMN gene dosage analysis. J Mol Diagn 2002; 4: 185–190.
Ogino S, Leonard DG, Rennert H, Gao S, Wilson RB : Heteroduplex formation in SMN gene dosage analysis. J Mol Diagn 2001; 3: 150–157.
Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B : Quantitative analyses of SMN1 and SMN2 based on real-time LightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 2002; 70: 358–368.
Corcia P, Mayeux-Portas V, Khoris J et al: Abnormal SMN1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann Neurol 2002; 51: 243–246.
Cusin V, Clermont O, Gerard B, Chantereau D, Elion J : Prevalence of SMN1 deletion and duplication in carrier and normal populations: implication for genetic counselling. J Med Genet 2003; 40: E39.
Anhuf D, Eggermann T, Rudnik-Schoneborn S, Zerres K : Determination of SMN1 and SMN2 copy number using TaqMan™ technology. Hum Mutat 2003; 22: 74–78.
Wirth B, Herz M, Wetter A et al: Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype–phenotype correlation, and implications for genetic counseling. Am J Hum Genet 1999; 64: 1340–1356.
Gerard B, Ginet N, Matthijs G et al: Genotype determination at the survival motor neuron locus in a normal population and SMA carriers using competitive PCR and primer extension. Hum Mutat 2000; 16: 253–263.
Mailman MD, Heinz JW, Papp AC et al: Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med 2002; 4: 20–26.
Vitali T, Sossi V, Tiziano F et al: Detection of the survival motor neuron (SMN) genes by FISH: further evidence for a role for SMN2 in the modulation of disease severity in SMA patients. Hum Mol Genet 1999; 8: 2525–2532.
Stephens M, Smith NJ, Donnelly P : A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989.
Brahe C, Servidei S, Zappata S et al: Genetic homogeneity between childhood-onset and adult-onset autosomal recessive spinal muscular atrophy. Lancet 1995; 346: 741–742.
Cobben JM, van der Steege G, Grootscholten P et al: Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am J Hum Genet 1995; 57: 805–808.
DiDonato CJ, Ingraham SE, Mendell JR et al: Deletion and conversion in spinal muscular atrophy patients: is there a relationship to severity? Ann Neurol 1997; 41: 230–237.
Hahnen E, Forkert R, Marke C et al: Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet 1995; 4: 1927–1933.
Somerville MJ, Hunter AG, Aubry HL et al: Clinical application of the molecular diagnosis of spinal muscular atrophy: deletions of neuronal apoptosis inhibitor protein and survival motor neuron genes. Am J Med Genet 1997; 69: 159–165.
Wang CH, Xu J, Carter TA et al: Characterization of survival motor neuron (SMNT) gene deletions in asymptomatic carriers of spinal muscular atrophy. Hum Mol Genet 1996; 5: 359–365.
Cusco II, Barcelo MJ, Baiget M, Tizzano EF : Implementation of SMA carrier testing in genetic laboratories: comparison of two methods for quantifying the SMN1 gene. Hum Mutat 2002; 20: 452–459.
Monani UR, Coovert DD, Burghes AH : Animal models of spinal muscular atrophy. Hum Mol Genet 2000; 9: 2451–2457.
Sabeti PC, Reich DE, Higgins JM et al: Detecting recent positive selection in the human genome from haplotype structure. Nature 2002; 419: 832–837.
Li WH, Ellsworth DL, Krushkal J, Chang BH, Hewett-Emmett D : Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol 1996; 5: 182–187.
Gibbons A : When it comes to evolution, humans are in the slow class. Science 1995; 267: 1907–1908.
Rochette CF, Gilbert N, Simard LR : SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet 2001; 108: 255–266.
Kondrashov AS : Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat 2003; 21: 12–27.
Nachman MW, Crowell SL : Estimate of the mutation rate per nucleotide in humans. Genetics 2000; 156: 297–304.
Acknowledgements
We thank H Rennert, M Dean and C O’hUigin for helpful discussions; and DGB Leonard, V Van Deerlin, and CS Fuchs for supporting various aspects of the work. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This work has been funded in whole or in part with Federal Funds from the National Cancer Institute and the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
(Supplementary information accompanies the paper on European Journal of Human Genetics website http://www.nature.com/ejhg).
Supplementary information
Rights and permissions
About this article
Cite this article
Ogino, S., Wilson, R. & Gold, B. New insights on the evolution of the SMN1 and SMN2 region: simulation and meta-analysis for allele and haplotype frequency calculations. Eur J Hum Genet 12, 1015–1023 (2004). https://doi.org/10.1038/sj.ejhg.5201288
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201288
Keywords
This article is cited by
-
Healthcare resource utilisation and direct medical cost for individuals with 5q spinal muscular atrophy in Sweden
The European Journal of Health Economics (2024)
-
Cost-Effectiveness of Newborn Screening for Spinal Muscular Atrophy in England
Neurology and Therapy (2023)
-
Spinal muscular atrophy
Nature Reviews Disease Primers (2022)
-
Cost-effectiveness analysis of gene-based therapies for patients with spinal muscular atrophy type I in Australia
Journal of Neurology (2022)
-
Costs of Illness of Spinal Muscular Atrophy: A Systematic Review
Applied Health Economics and Health Policy (2021)