Abstract
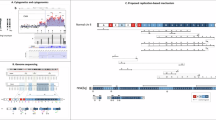
Duplicons, that is, DNA sequences with minimum length 10 kb and a high sequence similarity, are known to cause unequal homologous recombination, leading to deletions and the reciprocal duplications. In this study, we designed a Multiplex Amplifiable Probe Hybridisation (MAPH) assay containing 63 exon-specific single-copy sequences from within a selection of the 169 regions flanked by duplicons that were identified, at a first pass, in 2001. Subsequently, we determined the frequency of chromosomal rearrangements among patients with developmental delay (DD) and/or congenital malformations (CM). In addition, we tried to identify new regions involved in DD/CM using the same assay. In 105 patients, six imbalances (5.8%) were detected and verified. Three of these were located in microdeletion-related regions, two alterations were polymorphic duplications and the effect of the last alteration is currently unknown. The same study population was tested for rearrangements in regions with no known duplicons nearby, using a set of probes derived from 58 function-selected genes. The latter screening revealed two alterations. As expected, the alteration frequency per unit of DNA is much higher in regions flanked by duplicons (fraction of the genome tested: 5.2%) compared to regions without known duplicons nearby (fraction of the genome tested: 24.5–90.2%). We were able to detect three novel rearrangements, including the previously undescribed reciprocal duplication of the Williams Beuren critical region, a subduplicon alteration within this region and a duplication on chromosome band 16p13.11. Our results support the hypothesis that regions flanked by duplicons are enriched for copy number variations.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Leao JC, Bargman GJ, Neu RL, Kajii T, Gardner LI : New syndrome associated with partial deletion of short arms of chromosome No. 4. Clinical manifestations of hypospadias, beaked nose, abnormal iris, hemangioma of forehead, seizures, and other anomalies. J Am Med Assoc 1967; 202: 434–437.
Alfi O, Donnell GN, Crandall BF, Derencsenyi A, Menon R : Deletion of the short arm of chromosome no.9 (46,9p-): a new deletion syndrome. Ann Genet 1973; 16: 17–22.
Schinzel A, Auf der MP, Moser H : Partial deletion of long arm of chromosome 11[del(11)(q23)]: Jacobsen syndrome. Two new cases and review of the clinical findings. J Med Genet 1977; 14: 438–444.
Greenberg F, Crowder WE, Paschall V, Colon-Linares J, Lubianski B, Ledbetter DH : Familial DiGeorge syndrome and associated partial monosomy of chromosome 22. Hum Genet 1984; 65: 317–319.
Lupski JR, Wise CA, Kuwano A et al: Gene dosage is a mechanism for Charcot–Marie–Tooth disease type 1A. Nat Genet 1992; 1: 29–33.
Chance PF, Abbas N, Lensch MW et al: Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 1994; 3: 223–228.
Emanuel BS, Shaikh TH : Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat Rev Genet 2001; 2: 791–800.
Stankiewicz P, Lupski JR : Genome architecture, rearrangements and genomic disorders. Trends Genet 2002; 18: 74–82.
Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE : Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 2001; 11: 1005–1017.
Eichler EE : Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 2001; 17: 661–669.
Bailey JA, Gu Z, Clark RA et al: Recent segmental duplications in the human genome. Science 2002; 297: 1003–1007.
White S, Kalf M, Liu Q et al: Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet 2002; 71: 365–374.
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G : Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002; 30: e57.
White SJ, Vink GR, Kriek M et al: Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat 2004; 24: 86–92.
Dauwerse JG, Jumelet EA, Wessels JW et al: Extensive cross-homology between the long and short arm of chromosome 16 may explain leukemic inversions and translocations. Blood 1992; 79: 1299–1304.
Knijnenburg J, Szuhai K, Giltay J et al: Insights from genomic microarrays into structural chromosome rearrangements. Am J Med Genet A 2005; 132: 36–40.
Brewer C, Holloway S, Zawalnyski P, Schinzel A, Fitz Patrick D : A chromosomal duplication map of malformations: regions of suspected haplo- and triplolethality – and tolerance of segmental aneuploidy – in humans. Am J Hum Genet 1999; 64: 1702–1708.
Shaw CJ, Withers MA, Lupski JR : Uncommon deletions of the Smith–Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates. Am J Hum Genet 2004; 75: 75–81.
Bailey JA, Liu G, Eichler EE : An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 2003; 73: 823–834.
Lupski JR : Charcot–Marie–Tooth disease: lessons in genetic mechanisms. Mol Med 1998; 4: 3–11.
Shaw-Smith C, Redon R, Rickman L et al: Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet 2004; 41: 241–248.
Ensenauer RE, Adeyinka A, Flynn HC et al: Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet 2003; 73: 1027–1040.
Potocki L, Chen KS, Park SS et al: Molecular mechanism for duplication 17p11.2 – the homologous recombination reciprocal of the Smith–Magenis microdeletion. Nat Genet 2000; 24: 84–87.
Stankiewicz P, Lupski JR : Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 2002; 12: 312–319.
Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA : Mutational mechanisms of Williams–Beuren syndrome deletions. Am J Hum Genet 2003; 73: 131–151.
Peoples R, Franke Y, Wang Y et al: A physical map, including a BAC/PAC clone contig, of the Williams–Beuren syndrome–deletion region at 7q11.23. Am J Hum Genet 2000; 66: 47–68.
Shaw CJ, Bi W, Lupski JR : Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 2002; 71: 1072–1081.
Yan X, Li F, Liang Y et al: Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol Cell Biol 2003; 23: 1239–1250.
Sebat J, Lakshmi B, Troge J et al: Large-scale copy number polymorphism in the human genome. Science 2004; 305: 525–528.
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Sharp AJ, Locke DP, McGrath SD et al: Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 2005; 77: 78–88.
Acknowledgements
We thank Hans Dauwerse, Kerstin Hansson, Jeroen Nijhuis for the FISH analysis, Yvonne Hilhorst for providing clinical information, Peter de Knijff for parental marker analysis. MK is funded by Zon-Mw (AGIKO fellowship 940-37-032), SW is funded by ZonMw (nr 912-04-047).
Author information
Authors and Affiliations
Corresponding author
Additional information
Electronic-Database Information
† Leiden Muscular Dystrophy Pages, http://www.dmd.nl/DMD_MAPH.html
† Prophet (http://www.basic.nwu.edu/biotools/prophet.html)
† UCSC Genome Browser (http://genome.ucsc.edu/)
† Ensembl (http://www.ensembl.org)
† Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
† Genome Variation Database, http://projects.tcag.ca/variation/.
Note added in proof
While this work was under review, another patient was described (Severe expressive-language delay related to duplication of the Williams-Beuren locus, MJ Somerville et al. N Engl J Med 2005; 353:1694–1701, October 20, 2005) with a duplication of the WBS region. We have assessed the phenotype of our patient in the light of the reported clinical features (language deficiency but good spatial abilities). Considering the age of our patient, we could not assess the spatial abilities, but our patient did present with (moderate) language disability.
Rights and permissions
About this article
Cite this article
Kriek, M., White, S., Szuhai, K. et al. Copy number variation in regions flanked (or unflanked) by duplicons among patients with developmental delay and/or congenital malformations; detection of reciprocal and partial Williams-Beuren duplications. Eur J Hum Genet 14, 180–189 (2006). https://doi.org/10.1038/sj.ejhg.5201540
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201540
Keywords
This article is cited by
-
Evolution of the Cdk-activator Speedy/RINGO in vertebrates
Cellular and Molecular Life Sciences (2012)
-
Association of GTF2i in the Williams-Beuren Syndrome Critical Region with Autism Spectrum Disorders
Journal of Autism and Developmental Disorders (2012)
-
Phenotypic manifestations of copy number variation in chromosome 16p13.11
European Journal of Human Genetics (2011)
-
Copy number variants at Williams–Beuren syndrome 7q11.23 region
Human Genetics (2010)
-
Copy number variations (CNVs) identified in Korean individuals
BMC Genomics (2008)