Abstract
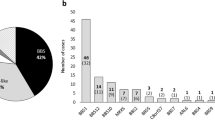
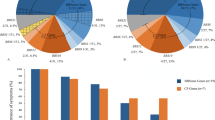
The extensive genetic heterogeneity of Bardet–Biedl syndrome (BBS) is documented by the identification, by classical linkage analysis complemented recently by comparative genomic approaches, of nine genes (BBS1–9) that account cumulatively for about 50% of patients. The BBS genes appear implicated in cilia and basal body assembly or function. In order to find new BBS genes, we performed SNP homozygosity mapping analysis in an extended consanguineous family living in a small Lebanese village. This uncovered an unexpectedly complex pattern of mutations, and led us to identify a novel BBS gene (BBS10). In one sibship of the pedigree, a BBS2 homozygous mutation was identified, while in three other sibships, a homozygous missense mutation was identified in a gene encoding a vertebrate-specific chaperonine-like protein (BBS10). The single patient in the last sibship was a compound heterozygote for the above BBS10 mutation and another one in the same gene. Although triallelism (three deleterious alleles in the same patient) has been described in some BBS families, we have to date no evidence that this is the case in the present family. The analysis of this family challenged linkage analysis based on the expectation of a single locus and mutation. The very high informativeness of SNP arrays was instrumental in elucidating this case, which illustrates possible pitfalls of homozygosity mapping in extended families, and that can be explained by the rather high prevalence of heterozygous carriers of BBS mutations (estimated at one in 50 in Europeans).
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Green JS, Parfrey PS, Harnett JD et al: The cardinal manifestations of Bardet–Biedl syndrome, a form of Laurence–Moon–Biedl. N Engl J Med 1989; 321: 1002–1009.
Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA : New criteria for improved diagnosis of Bardet–Biedl syndrome: results of a population survey. J Med Genet 1999; 36: 437–446.
Mykytyn K, Nishimura DY, Searby CC et al: Identification of the gene (BBS1) most commonly involved in Bardet–Bield syndrome, a complex human obesity syndrome. Nat Genet 2002; 31: 435–438.
Nishimura DY, Searby CC, Carmi R et al: Positional cloning of a novel gene on chromosome 16q causing Bardet–Biedl syndrome. Hum Mol Genet 2001; 10: 865–874.
Fan Y, Esmail MA, Ansley SJ et al: Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet–Biedl syndrome. Nat Genet 2004; 36: 989–993.
Chiang AP, Nishimura D, Searby C et al: Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet–Biedl syndrome (BBS3). Am J Hum Genet 2004; 75: 475–484.
Mykytyn K, Braun T, Carmi R et al: Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 2001; 28: 188–191.
Li JB, Gerdes JM, Haycraft CJ et al: Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 2004; 117: 541–552.
Slavotinek AM, Stone EM, Mykytyn K et al: Mutations in MKKS cause Bardet–Biedl syndrome. Nat Genet 2000; 26: 15–16.
Katsanis N, Beales PL, Woods MO et al: Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet–Biedl syndrome. Nat Genet 2000; 26: 67–70.
Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, Katsanis N : Identification of a novel Bardet–Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet 2003; 72: 650–658.
Ansley SJ, Badano JL, Blacque OE et al: Basal body dysfunction is a likely cause of pleiotropic Bardet–Biedl syndrome. Nature 2003; 425: 628–633.
Nishimura DY, Swiderski RE, Searby CC et al: Comparative genomics and gene expression analysis identifies BBS9, a new Bardet–Biedl syndrome gene. Am J Hum genet 2005; 77: 1021–1033.
Katsanis N, Ansley SJ, Badano JL et al: Triallelic inheritance in Bardet–Biedl syndrome, a Mendelian recessive disorder. Science 2001; 293: 2256–2259.
Badano JL, Kim JC, Hoskins BE et al: Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet–Biedl patients with two mutations at a second BBS locus. Hum Mol Genet 2003; 12: 1651–1659.
Fauser S, Munz M, Besch D : Further support for digenic inheritance in Bardet–Biedl syndrome. J Med Genet 2003; 40: e104.
Beales PL, Badano JL, Ross AJ et al: Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-mendelian Bardet–Biedl syndrome. Am J Hum Genet 2003; 72: 1187–1199.
Katsanis N : The oligogenic properties of Bardet–Biedl syndrome. Hum Mol genet 2004; Spec. No. 1: R65–R71.
Mykytyn K, Nishimura DY, Searby CC et al: Evaluation of complex inheritance involving the most common Bardet–Biedl syndrome locus (BBS1). Am J Hum Genet 2003; 72: 429–437, (E-pub ahead of print Jan 10 2003).
Moore SJ, Green JS, Fan Y et al: Clinical and genetic epidemiology of Bardet–Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. Am J Med Genet A 2005; 132: 352–360.
Hichri H, Stoetzel C, Laurier V et al: Testing for triallelism: analysis of six BBS genes in a Bardet–Biedl syndrome family cohort. Eur J Hum Genet 2005; 13: 607–616.
Nakane T, Biesecker LG : No evidence for triallelic inheritance of MKKS/BBS loci in Amish Mckusick–Kaufman syndrome. Am J Med Genet A 2005; 138: 32–34.
Ross AJ, May-Simera H, Eichers ER et al: Disruption of Bardet–Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet 2005; 37: 1135–1140.
Stoetzel C, Laurier V, Davis E et al: A novel gene encoding a vertebrate-specific chaperonin-like is a major BBS locus. Nat Genet 2006; 38: 521–524.
Miano MG, Jacobson SG, Carothers A et al: Pitfalls in homozygosity mapping. Am J Hum Genet 2000; 67: 1348–1351.
Lander ES, Botstein D : Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 1987; 236: 1567–1570.
Weissenbach J, Gyapay G, Dib C et al: A second-generation linkage map of the human genome. Nature 1992; 359: 794–801.
Ben Hamida C, Doerflinger N, Belal S et al: Localization of Friedreich ataxia phenotype with selective vitamin E deficiency to chromosome 8q by homozygosity mapping. Nat Genet 1993; 5: 195–200.
Pollak MR, Chou YH, Cerda JJ, Steinmann B et al: Homozygosity mapping of the gene for alkaptonuria to chromosome 3q2. Nat Genet 1993; 5: 201–204.
Kwitek-Black AE, Carmi R, Duyk GM et al: linkage of Bardet–Biedl syndrome to chromosome 16q and evidence for non-allelic genetic heterogeneity. Nat Genet 1993; 5: 392–396.
Janecke AR, Thompson DA, Utermann G et al: Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet 2004; 36: 850–854.
Leppert M, Baird L, Anderson KL, Otterud B, Lupski JR, Lewis RA : Bardet–Biedl syndrome is linked to DNA markers on chromosome 11q and is genetically heterogeneous. Nat Genet 1994; 7: 108–112.
Sheffield VC, Carmi R, Kwitek-Black A et al: Identification of a Bardet–Biedl syndrome locus on chromosome 3 and evaluation of an efficient approach to homozygosity mapping. Hum Mol Genet 1994; 3: 1331–1335.
Bruford EA, Riise R, Teague PW et al: Linkage mapping in 29 Bardet–Biedl syndrome families confirms loci in chromosomal regions 11q13, 15q22–3, and 16q21. Genomics 1997; 41: 93–99.
Young T-L, Penney L, Woods MO et al: A fifth locus for Bardet–Biedl syndrome maps to chromosome 2q31. Am J Hum Genet 1999; 64: 900–904.
Gibson J, Morton NE, Collins A : Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 2006; 15: 789–795.
Pannain S, Weiss RE, Jackson CE et al: Two different mutations in the thyroid peroxidase gene of a large inbred Amish kindred: power and limits of homozygosity mapping. J Clin Endocrinol Metab 1999; 84: 1061–1071.
Allamand V, Broux O, Richard I et al: Preferential localization of the limb–girdle muscular dystrophy type 2A gene in the proximal part of a 1-cM 15q15.1–q15.3 interval. Am J Hum Genet 1995; 56: 1417–1430.
Richard I, Broux O, Allamand V et al: Mutations in the proteolytic enzyme calpain 3 cause limb–girdle muscular dystrophy type 2A. Cell 1995; 81: 27–40.
Beales PL, Warner AM, Hitman GA, Thakker R, Flinter FA : Bardet–Biedl syndrome: a molecular and phenotypic study of 18 families. J Med Genet 1997; 34: 92–98.
Klein D, Ammann F : The syndrome of Laurence—Moon–Bardet–Biedl and allied diseases in Switzerland. Clinical, genetic and epidemiological studies. J Neurol Sci 1969; 9: 479–513.
Farag TI, Teebi AS : High incidence of Bardet–Biedl syndrome among the Bedouin. Clin Genet 1989; 36: 463–464.
Green JS, Harnett JD, Farid NR et al: The cardinal manifestations of Bardet–Biedl syndrome, a form of Laurence–Moon–Biedl syndrome. N Engl J Med 1989; 321: 1002–1009.
Fan Y, Rahman P, Peddle L et al: Bardet–Biedl syndrome 1 genotype and obesity in the Newfoundland population. Int J Obes Relat Metab Disord 2004; 28: 680–684.
Andersen KL, Echwald SM, Larsen LH et al: Variation of the McKusick–Kaufman gene and studies of relationships with common forms of obesity. J Clin Endocrinol Metab 2005; 90: 225–230.
Badano JL, Leitch CC, Ansley SJ et al: Dissection of epistasis in oligogenic Bardet–Biedl syndrome. Nature 2006; 439: 326–330.
Avidor-Reiss T, Maer AM, Koundakjian E et al: Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 2004; 117: 527–539.
Chiang AP, Beck JS, Yen HJ et al: Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci USA 2006; 103: 6287–6292.
Acknowledgements
We acknowledge the financial support of: Ministère de la Recherche (PHRC national 2002), RETINA France, LION's club du Kochersberg and la Fédération des Maladies Orphelines. We also thank the PNRV–INSERM programme (Programme National sur la Recherche pour la Vision). We thank the constant technical help of the IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch-Graffenstaden), especially Catherine Grussenmeyer (Microarray department). We acknowledge the INSERM, CNRS and Université Louis Pasteur. Jean Muller was supported by grants from the National Research Fund and the MCESR of Luxembourg.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Laurier, V., Stoetzel, C., Muller, J. et al. Pitfalls of homozygosity mapping: an extended consanguineous Bardet–Biedl syndrome family with two mutant genes (BBS2, BBS10), three mutations, but no triallelism. Eur J Hum Genet 14, 1195–1203 (2006). https://doi.org/10.1038/sj.ejhg.5201688
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201688
Keywords
This article is cited by
-
Novel biallelic splice-site BBS1 variants in Bardet–Biedle syndrome: a case report of the first Japanese patient
Documenta Ophthalmologica (2020)
-
Whole genome sequencing identifies a novel ALMS1 gene mutation in two Chinese siblings with Alström syndrome
BMC Medical Genetics (2017)
-
Genetic and clinical characterization of Pakistani families with Bardet-Biedl syndrome extends the genetic and phenotypic spectrum
Scientific Reports (2016)
-
Characterization of CCDC28B reveals its role in ciliogenesis and provides insight to understand its modifier effect on Bardet–Biedl syndrome
Human Genetics (2013)
-
Phenotypic variability of Bardet-Biedl syndrome: focusing on the kidney
Pediatric Nephrology (2012)