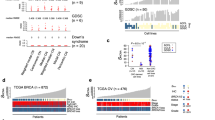
Abstract
Array-based genome-wide screening methods were recently introduced to clinical practice in order to detect small genomic imbalances that may cause severe genetic disorders. The continuous advancement of such methods plays an extremely important role in diagnostic genetics and medical genomics. We have modified and adapted the original multiplex amplifiable probe hybridization (MAPH) to a novel microarray format providing an important new diagnostic tool for detection of small size copy-number changes in any locus of human genome. Here, we describe the new array-MAPH diagnostic method and show proof of concept through fabrication, interrogation and validation of a human chromosome X-specific array. We have developed new bioinformatic tools and methodology for designing and producing amplifiable hybridization probes (200–600 bp) for array-MAPH. We designed 558 chromosome X-specific probes with median spacing 238 kb and 107 autosomal probes, which were spotted onto microarrays. DNA samples from normal individuals and patients with known and unknown chromosome X aberrations were analyzed for validation. Array-MAPH detected exactly the same deletions and duplications in blind studies, as well as other unknown small size deletions showing its accuracy and sensitivity. All results were confirmed by fluorescence in situ hybridization and probe-specific PCR. Array-MAPH is a new microarray-based diagnostic tool for the detection of small-scale copy-number changes in complex genomes, which may be useful for genotype–phenotype correlations, identification of new genes, studying genetic variation and provision of genetic services.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Test and Technology Transfer Committee: Technical and clinical assessment of fluorescence in situ hybridization: an ACMG/ASHG position statement. I. Technical considerations. Genet Med 2000; 2: 356–361.
Patsalis PC, Evangelidou P, Charalambous S, Sismani C : Fluorescence in situ hybridization characterization of apparently balanced translocation reveals cryptic complex chromosomal rearrangements with unexpected level of complexity. Eur J Hum Genet 2004; 12: 647–653.
Knight SJ, Flint J : Perfect endings: a review of subtelomeric probes and their use in clinical diagnosis. J Med Genet 2000; 37: 401–409.
Kallioniemi A, Kallioniemi OP, Sudar D et al: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992; 258: 818–821.
Pinkel D, Segraves R, Sudar D et al: High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 1998; 20: 207–211.
Pollack JR, Perou CM, Alizadeh AA et al: Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 1999; 23: 41–46.
Solinas-Toldo S, Lampel S, Stilgenbauer S et al: Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer 1997; 20: 399–407.
Snijders AM, Nowak N, Segraves R et al: Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 2001; 29: 263–264.
Lucito R, West J, Reiner A et al: Detecting gene copy number fluctuations in tumor cells by microarray analysis of genomic representations. Genome Res 2000; 10: 1726–1736.
Lucito R, Healy J, Alexander J et al: Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res 2003; 13: 2291–2305.
Zhao X, Li C, Paez JG et al: An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res 2004; 64: 3060–3071.
Huang J, Wei W, Zhang J et al: Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genomics 2004; 1: 287–299.
Selzer RR, Richmond TA, Pofahl NJ et al: Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 2005; 44: 305–319.
Ishkanian AS, Malloff CA, Watson SK et al: A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet 2004; 36: 299–303.
Armour JA, Sismani C, Patsalis PC, Cross G : Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res 2000; 28: 605–609.
Hollox EJ, Akrami SM, Armour JA : DNA copy number analysis by MAPH: molecular diagnostic applications. Expert Rev Mol Diagn 2002; 2: 370–378.
Patsalis PC, Kousoulidou L, Sismani C, Mannik K, Kurg A : MAPH: from gels to microarrays. Eur J Med Genet 2005; 48: 241–249.
Altschul SF, Madden TL, Schaffer AA et al: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–3402.
Rozen S, Skaletsky H : Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000; 132: 365–386.
Cai WW, Mao JH, Chow CW, Damani S, Balmain A, Bradley A : Genome-wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat Biotechnol 2002; 20: 393–396.
Barrett MT, Scheffer A, Ben-Dor A et al: Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc Natl Acad Sci USA 2004; 101: 17765–17770.
Mantripragada KK, Buckley PG, Jarbo C, Menzel U, Dumanski JP : Development of NF2 gene specific, strictly sequence defined diagnostic microarray for deletion detection. J Mol Med 2003; 81: 443–451.
Mantripragada KK, Thuresson AC, Piotrowski A et al: Identification of novel deletion breakpoints bordered by segmental duplications in the NF1 locus using high resolution array-CGH. J Med Genet 2006; 43: 28–38.
Mantripragada KK, Tapia-Paez I, Blennow E, Nilsson P, Wedell A, Dumanski JP : DNA copy-number analysis of the 22q11 deletion-syndrome region using array-CGH with genomic and PCR-based targets. Int J Mol Med 2004; 13: 273–279.
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Sebat J, Lakshmi B, Troge J et al: Large-scale copy number polymorphism in the human genome. Science 2004; 305: 525–528.
Lucito R, Nakimura M, West JA et al: Genetic analysis using genomic representations. Proc Natl Acad Sci USA 1998; 95: 4487–4492.
Kennedy GC, Matsuzaki H, Dong S et al: Large-scale genotyping of complex DNA. Nat Biotechnol 2003; 21: 1233–1237.
Acknowledgements
This work was funded by the grants 30/2001 from the Cyprus RPF, QLRT-2001-01810 from the EURO-MRX EU, 5467 from the Estonian Science Foundation, by 0182649s04 from the Estonian Ministry of Education and Research and by 070191/Z/03/Z from the Wellcome Trust International Senior Research Grant. We thank D Andreou, C Tryfonos, E Hadjiyanni, C Pitta, C Antoniades, S Bashiardes and G Slavin for their contribution. Many thanks to Professor J Vermeesch, Dr K Õunap and Dr R Žordania for the provision of DNA samples and the Wellcome Trust Sanger Institute for the provision of BAC/PAC clones.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Patsalis, P., Kousoulidou, L., Männik, K. et al. Detection of small genomic imbalances using microarray-based multiplex amplifiable probe hybridization. Eur J Hum Genet 15, 162–172 (2007). https://doi.org/10.1038/sj.ejhg.5201738
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201738
Keywords
This article is cited by
-
Progress in the detection of human genome structural variations
Science in China Series C: Life Sciences (2009)
-
Array-MAPH: a methodology for the detection of locus copy-number changes in complex genomes
Nature Protocols (2008)