Abstract
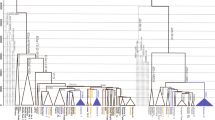
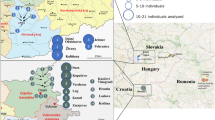
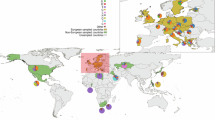
The island of Crete, credited by some historical scholars as a central crucible of western civilization, has been under continuous archeological investigation since the second half of the nineteenth century. In the present work, the geographic stratification of the contemporary Cretan Y-chromosome gene pool was assessed by high-resolution haplotyping to investigate the potential imprints of past colonization episodes and the population substructure. In addition to analyzing the possible geographic origins of Y-chromosome lineages in relatively accessible areas of the island, this study includes samples from the isolated interior of the Lasithi Plateau – a mountain plain located in eastern Crete. The potential significance of the results from the latter region is underscored by the possibility that this region was used as a Minoan refugium. Comparisons of Y-haplogroup frequencies among three Cretan populations as well as with published data from additional Mediterranean locations revealed significant differences in the frequency distributions of Y-chromosome haplogroups within the island. The most outstanding differences were observed in haplogroups J2 and R1, with the predominance of haplogroup R lineages in the Lasithi Plateau and of haplogroup J lineages in the more accessible regions of the island. Y-STR-based analyses demonstrated the close affinity that R1a1 chromosomes from the Lasithi Plateau shared with those from the Balkans, but not with those from lowland eastern Crete. In contrast, Cretan R1b microsatellite-defined haplotypes displayed more resemblance to those from Northeast Italy than to those from Turkey and the Balkans.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Broodbank C, Strasser T : Migrant farmers and the Neolithic colonisation of Crete. Antiquity 1991; 65: 233–245.
Fitton JL : Minoans. London: The British Museum Press, 2002.
Nowicki K : Defensible sites in Crete c1200 – 800 BC (LMIIIB/IIIC through Early Geometric). Université de Liège, Liège, University of Texas, Austin: Aegaeum Series 21, 2000.
Watrous LV : Lasithi: a history of settlement on a highland plain in Crete. Hesperia Supplement 1982; 18: i-iii+v+vii-xiv+1-89+91-101+103-122.
Rackham O, Moody J : The making of the Cretan landscape. Manchester: Manchester University Press, 1996.
Di Giacomo F, Luca F, Anagnou N et al: Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects. Mol Phylogenet Evol 2003; 28: 387–395.
Malaspina P, Tsopanomichalou M, Duman T et al: A multistep process for the dispersal of a Y chromosomal lineage in the Mediterranean area. Ann Hum Genet 2001; 65: 339–349.
Sambrook J, Russell DW : Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press, 2001.
Martinez L, Reategui EP, Fonseca LR et al: Superimposing polymorphism: the case of a point mutation within a polymorphic Alu insertion. Hum Hered 2005; 59: 109–117.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ : Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered 2006; 61: 132–143.
Nei M, Tajima F : DNA polymorphism detectable by restriction endonucleases. Genet 1981; 97: 145–163.
Rohlf F : NTSYSpc. Setauket, New York: Exeter Publishing, 2002.
Cinnioglu C, King R, Kivisild T et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Zhivotovsky LA : Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol 2001; 18: 700–709.
Zhivotovsky LA, Underhill PA, Cinnioglu C et al: The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 2004; 74: 50–61.
Perièiæ M, Lauc LB, Klaric IM et al: High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations. Mol Biol Evol 2005; 22: 1964–1975.
Wilson I, Weale M, Balding D : BATWING: Bayesian analysis of trees with internal node generation. Aberdeen: Department of Mathematical Sciences, University of Aberdeen, 2000.
Rohl A : Network 2.0: a program package for calculating phylogenetic networks. Mathematisches Seminar, Hamburg, Germany: University of Hamburg, 1997.
Cruciani F, La Fratta R, Santolamazza P et al: Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet 2004; 74: 1014–1022.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 2000; 290: 1155–1159.
Francalacci P, Morelli L, Underhill PA et al: Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am J Phys Anthropol 2003; 121: 270–279.
Arredi B, Poloni ES, Paracchini S et al: A predominantly Neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet 2004; 75: 338–345.
Luis JR, Rowold DJ, Regueiro M et al: The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 2004; 74: 532–544.
Shen P, Lavi T, Kivisild T et al: Reconstruction of patrilineages and matrilineages of Samaritans and other Israeli populations from Y-chromosome and mitochondrial DNA sequence variation. Hum Mutat 2004; 24: 248–260.
Di Giacomo F, Luca F, Popa LO et al: Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. Hum Genet 2004; 115: 357–371.
Semino O, Magri C, Benuzzi G et al: Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004; 74: 1023–1034.
Hammer MF, Karafet T, Rasanayagam A et al: Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 1998; 15: 427–441.
Cruciani F, La Fratta R, Torroni A, Underhill PA, Scozzari R : Molecular dissection of the Y chromosome haplogroup E-M78 (E3b1a): A posteriori evaluation of a microsatellite-network-based approach through six new biallelic markers. Hum Mutat 2006; 27: 831–832.
Malaspina P, Cruciani F, Santolamazza P et al: Patterns of male-specific inter-population divergence in Europe, West Asia and North Africa. Ann Hum Genet 2000; 64: 395–412.
Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249.
Marjanovic D, Fornarino S, Montagna S et al: The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups. Ann Hum Genet 2005; 69: 757–763.
Acknowledgements
We gratefully acknowledge Dr Manuela M Regueiro for her technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Martinez, L., Underhill, P., Zhivotovsky, L. et al. Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau. Eur J Hum Genet 15, 485–493 (2007). https://doi.org/10.1038/sj.ejhg.5201769
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201769
Keywords
This article is cited by
-
The Y chromosome of autochthonous Basque populations and the Bronze Age replacement
Scientific Reports (2021)
-
The Greeks in the West: genetic signatures of the Hellenic colonisation in southern Italy and Sicily
European Journal of Human Genetics (2016)
-
Ladakh, India: the land of high passes and genetic heterogeneity reveals a confluence of migrations
European Journal of Human Genetics (2016)
-
The phylogenetic and geographic structure of Y-chromosome haplogroup R1a
European Journal of Human Genetics (2015)
-
Genetic characterization of Greek population isolates reveals strong genetic drift at missense and trait-associated variants
Nature Communications (2014)