Abstract
Aim:
hERG potassium channels display miscellaneous interactions with diverse chemical scaffolds. In this study we assessed the hERG inhibition in a large compound library of diverse chemical entities and provided data for better understanding of the mechanisms underlying promiscuity of hERG inhibition.
Methods:
Approximately 300 000 compounds contained in Molecular Library Small Molecular Repository (MLSMR) library were tested. Compound profiling was conducted on hERG-CHO cells using the automated patch-clamp platform–IonWorks Quattro™.
Results:
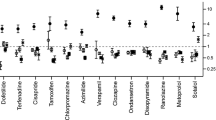
The compound library was tested at 1 and 10 μmol/L. IC50 values were predicted using a modified 4-parameter logistic model. Inhibitor hits were binned into three groups based on their potency: high (IC50<1 μmol/L), intermediate (1 μmol/L< IC50<10 μmol/L), and low (IC50>10 μmol/L) with hit rates of 1.64%, 9.17% and 16.63%, respectively. Six physiochemical properties of each compound were acquired and calculated using ACD software to evaluate the correlation between hERG inhibition and the properties: hERG inhibition was positively correlative to the physiochemical properties ALogP, molecular weight and RTB, and negatively correlative to TPSA.
Conclusion:
Based on a large diverse compound collection, this study provides experimental evidence to understand the promiscuity of hERG inhibition. This study further demonstrates that hERG liability compounds tend to be more hydrophobic, high-molecular, flexible and polarizable.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Fermini B, Fossa AA . The impact of drug-induced QT interval prolongation on drug discovery and development. Nat Rev Drug Discov 2003; 2: 439–47.
Sanguinetti MC, Jiang C, Curran ME, Keating MT . A mechanistic link between an inherited and an acquired cardiac arrhythmia: hERG encodes the IKr potassium channel. Cell 1995; 81: 299–307.
Zhang KP, Yang BF, Li BX . Translational toxicology and rescue strategies of the hERG channel dysfunction: biochemical and molecular mechanistic aspects. Acta Pharmacol Sin 2014; 35: 1473–84.
Babcock JJ, Li M . hERG channel function: beyond long QT. Acta Pharmacol Sin 2013; 34: 329–35.
ICH S7B. Guideline on safety pharmacology studies for assessing the potential for delayed ventricular repolarization (QT interval prolongation) by human pharmaceuticals 2005.
Witchel HJ, Milnes JT, Mitcheson JS, Hancox JC . Troubleshooting problems with in vitro screening of drugs for QT interval prolongation using HERG K+ channels expressed in mammalian cell lines and Xenopus oocytes. J Pharmacol Toxicol Methods 2002; 48: 65–80.
Zou B, Yu H, Babcock JJ, Chanda P, Bader JS, McManus OB, et al. Profiling diverse compounds by flux- and electrophysiology-based primary screens for inhibition of human Ether-a-go-go related gene potassium channels. Assay Drug Dev Technol 2010; 8: 743–54.
Schmalhofer WA, Swensen AM, Thomas BS, Felix JP, Haedo RJ, Solly K, et al. A pharmacologically validated, high-capacity, functional thallium flux assay for the human Ether-a-go-go related gene potassium channel. Assay Drug Dev Technol 2010; 8: 714–26.
Mattmann ME, Yu H, Lin Z, Xu K, Huang X, Long S, et al. Identification of (R)-N-(4-(4-methoxyphenyl)thiazol-2-yl)-1-tosylpiperidine-2-carboxamide, ML277, as a novel, potent and selective Kv7.1 (KCNQ1) potassium channel activator. Bioorg Med Chem Lett 2012; 22: 5936–41.
Cheung YY, Yu H, Xu K, Zou B, Wu M, McManus OB, et al. Discovery of a series of 2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)acetamides as novel molecular switches that modulate modes of Kv7.2 (KCNQ2) channel pharmacology: identification of (S)-2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)butanamide (ML252) as a potent, brain penetrant Kv7.2 channel inhibitor. J Med Chem 2012; 55: 6975–9.
Yu H, Wu M, Townsend SD, Zou B, Long S, Daniels JS, et al. Discovery, synthesis, and structure activity relationship of a series of N-Aryl- bicyclo[2.2.1]heptane-2-carboxamides: Characterization of ML213 as a novel KCNQ2 and KCNQ4 potassium channel opener. ACS Chem Neurosci 2011; 2: 572–7.
Song M, Clark M . Development and evaluation of an in silico model for hERG binding. J Chem Inf Model 2006; 46: 392–400.
Doddareddy MR, Klaasse EC, Shagufta, Ijzerman AP, Bender A . Prospective validation of a comprehensive in silico hERG model and its applications to commercial compound and drug databases. ChemMedChem 2010; 5: 716–29.
Buturak B, Durdagi S, Noskov SY, Ildeniz AT . Designing of multi-targeted molecules using combination of molecular screening and in silico drug cardiotoxicity prediction approaches. J Mol Graph Model 2014; 50: 16–34.
Bidault Y . A flexible approach for optimising in silico ADME/Tox characterisation of lead candidates. Expert Opin Drug Metab Toxicol 2006; 2: 157–68.
Beresford AP, Segall M, Tarbit MH . In silico prediction of ADME properties: are we making progress? Curr Opin Drug Discov Devel 2004; 7: 36–42.
Beattie KA, Luscombe C, Williams G, Munoz-Muriedas J, Gavaghan DJ, Cui Y, et al. Evaluation of an in silico cardiac safety assay: using ion channel screening data to predict QT interval changes in the rabbit ventricular wedge. J Pharmacol Toxicol Methods 2013; 68: 88–96.
Liu LL, Lu J, Lu Y, Zheng MY, Luo XM, Zhu WL, et al. Novel Bayesian classification models for predicting compounds blocking hERG potassium channels. Acta Pharmacol Sin 2014; 35: 1093–102.
Schreiber SL, Kotz JD, Li M, Aube J, Austin CP, Reed JC, et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell 2015; 161: 1252–65.
Du F, Yu H, Zou B, Babcock J, Long S, Li M . hERGCentral: a large database to store, retrieve, and analyze compound-human Ether-a-go-go related gene channel interactions to facilitate cardiotoxicity assessment in drug development. Assay Drug Dev Technol 2011; 9: 580–8.
Du F, Babcock JJ, Yu H, Zou B, Li M . Global analysis reveals families of chemical motifs enriched for hERG inhibitors. PLoS One 2015; 10: e0118324.
Wang S, Li Y, Wang J, Chen L, Zhang L, Yu H, et al. ADMET evaluation in drug discovery. 12. Development of binary classification models for prediction of hERG potassium channel blockage. Mol Pharm 2012; 9: 996–1010.
Weiss JN . The Hill equation revisited: uses and misuses. FASEB J 1997; 11: 835–41.
Gadagkar SR, Call GB . Computational tools for fitting the Hill equation to dose-response curves. J Pharmacol Toxicol Methods 2015; 71: 68–76.
Krippendorff BF, Lienau P, Reichel A, Huisinga W . Optimizing classification of drug-drug interaction potential for CYP450 isoenzyme inhibition assays in early drug discovery. J Biomol Screen 2007; 12: 92–9.
Bowlby MR, Peri R, Zhang H, Dunlop J . hERG (KCNH2 or Kv11.1) K+ channels: screening for cardiac arrhythmia risk. Curr Drug Metab 2008; 9: 965–70.
Hernandez-Covarrubias C, Vilchis-Reyes MA, Yepez-Mulia L, Sanchez-Diaz R, Navarrete-Vazquez G, Hernandez-Campos A, et al. Exploring the interplay of physicochemical properties, membrane permeability and giardicidal activity of some benzimidazole derivatives. Eur J Med Chem 2012; 52: 193–204.
Manchester J, Walkup G, Rivin O, You Z . Evaluation of pKa estimation methods on 211 druglike compounds. J Chem Inf Model 2010; 50: 565–71.
Mo ZL, Faxel T, Yang YS, Gallavan R, Messing D, Bahinski A . Effect of compound plate composition on measurement of hERG current IC50 using PatchXpress. J Pharmacol Toxicol Methods 2009; 60: 39–44.
Pearlstein RA, Vaz RJ, Kang J, Chen XL, Preobrazhenskaya M, Shchekotikhin AE, et al. Characterization of hERG potassium channel inhibition using CoMSiA 3D QSAR and homology modeling approaches. Bioorg Med Chem Lett 2003; 13: 1829–35.
Gao F, Johnson DL, Ekins S, Janiszewski J, Kelly KG, Meyer RD, et al. Optimizing higher throughput methods to assess drug-drug interactions for CYP1A2, CYP2C9, CYP2C19, CYP2D6, rCYP2D6, and CYP3A4 in vitro using a single point IC50 . J Biomol Screen 2002; 7: 373–82.
Sebaugh JL . Guidelines for accurate EC50/IC50 estimation. Pharm Stat 2011; 10: 128–34.
Raschi E, Ceccarini L, De Ponti F, Recanatini M . hERG-related drug toxicity and models for predicting hERG liability and QT prolongation. Expert Opin Drug Metab Toxicol 2009; 5: 1005–21.
Titus SA, Beacham D, Shahane SA, Southall N, Xia M, Huang R, et al. A new homogeneous high-throughput screening assay for profiling compound activity on the human ether-a-go-go-related gene channel. Anal Biochem 2009; 394: 30–8.
Ding M, Stjernborg L, Albertson N . Application of cryopreserved cells to hERG screening using a non-radioactive Rb+ efflux assay. Assay Drug Dev Technol 2006; 4: 83–8.
Cheng CS, Alderman D, Kwash J, Dessaint J, Patel R, Lescoe MK, et al. A high-throughput HERG potassium channel function assay: an old assay with a new look. Drug Dev Ind Pharm 2002; 28: 177–91.
Rezazadeh S, Hesketh JC, Fedida D . Rb+ flux through hERG channels affects the potency of channel blocking drugs: correlation with data obtained using a high-throughput Rb+ efflux assay. J Biomol Screen 2004; 9: 588–97.
Tang W, Kang J, Wu X, Rampe D, Wang L, Shen H, et al. Development and evaluation of high throughput functional assay methods for hERG potassium channel. J Biomol Screen 2001; 6: 325–31.
Bridgland-Taylor MH, Hargreaves AC, Easter A, Orme A, Henthorn DC, Ding M, et al. Optimisation and validation of a medium-throughput electrophysiology-based hERG assay using IonWorks HT. J Pharmacol Toxicol Methods 2006; 54: 189–99.
Murphy SM, Palmer M, Poole MF, Padegimas L, Hunady K, Danzig J, et al. Evaluation of functional and binding assays in cells expressing either recombinant or endogenous hERG channel. J Pharmacol Toxicol Methods 2006; 54: 42–55.
Aptula AO, Cronin MT . Prediction of hERG K+ blocking potency: application of structural knowledge. SAR QSAR Environ Res 2004; 15: 399–411.
Acknowledgements
This work was supported by the grant from the National Institutes of Health of USA (U54 MH084691) (to Min LI), and National Natural Science Foundation of China (Grant 81470163 and 81503056) (to Hai-bo YU).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Tables are available at Acta Pharmacologica Sinica's website.
Supplementary information
Supplementary Information, Table S1
The well-characterized hERG inhibitors were validated by automated patch clamp-IonWorks Quattro™. (DOC 77 kb)
Supplementary Information, Table S2
Reported hERG inhibitors were validated by automated patch clamp-IonWorks Quattro™ (DOC 529 kb)
Rights and permissions
About this article
Cite this article
Yu, Hb., Zou, By., Wang, Xl. et al. Investigation of miscellaneous hERG inhibition in large diverse compound collection using automated patch-clamp assay. Acta Pharmacol Sin 37, 111–123 (2016). https://doi.org/10.1038/aps.2015.143
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/aps.2015.143
Keywords
This article is cited by
-
A structure-based computational workflow to predict liability and binding modes of small molecules to hERG
Scientific Reports (2020)
-
The High Cost of Stroke and Stroke Cytoprotection Research
Translational Stroke Research (2017)
-
Ion channels research in the post-genomic era
Acta Pharmacologica Sinica (2016)
-
Compilation and physicochemical classification analysis of a diverse hERG inhibition database
Journal of Computer-Aided Molecular Design (2016)