Abstract
TGF-beta1 has been shown to induce autophagy in certain cells but whether and how this action is exerted in muscle and whether this activity relates to TGF-beta1 control of muscle cell differentiation remains unknown. Here, we show that expression of the autophagy-promoting protein phosphoprotein enriched in diabetes/phosphoprotein enriched in astrocytes (PED/PEA-15) progressively declines during L6 and C2C12 skeletal muscle cell differentiation. PED/PEA-15 underwent rapid induction upon TGF-beta1 exposure of L6 and C2C12 myoblasts, accompanied by impaired differentiation into mature myotubes. TGF-beta1 also induced autophagy in the L6 and C2C12 cells through a PP2A/FoxO1-mediated mechanism. Both the TGF-beta1 effect on differentiation and that on autophagy were blocked by specific PED/PEA-15 ShRNAs. Myoblasts stably overexpressing PED/PEA-15 did not differentiate and showed markedly enhanced autophagy. In these same cells, the autophagy inhibitor 3-methyladenine rescued TGF-beta1 effect on both autophagy and myogenesis, indicating that PED/PEA-15 mediates TGF-beta1 effects in muscle. Muscles from transgenic mice overexpressing PED/PEA-15 featured a significant number of atrophic fibers, accompanied by increased light chain 3 (LC3)II to LC3I ratio and reduced PP2A/FoxO1 phosphorylation. Interestingly, these mice showed significantly impaired locomotor activity compared with their non-transgenic littermates. TGF-beta1 causes transcriptional upregulation of the autophagy-promoting gene PED/PEA-15, which in turn is capable to induce atrophic responses in skeletal muscle in vivo.
Similar content being viewed by others
Main
The superfamily of transforming growth factor-beta (TGF-beta) cytokines are ubiquitously expressed and induce a wide variety of effects ultimately depending on the cell context and including regulation of cell growth, proliferation, differentiation, adhesion, migration and apoptosis.1, 2, 3, 4, 5, 6 Also, different members of the TGF-beta superfamily, including the archetypal TGF-beta family member TGF-beta1, have been found to inhibit myogenic differentiation in primary cultures and in myoblast cell lines from different species.7, 8, 9 These findings have evoked considerable translational interest, as inhibitors of the TGF-beta signaling cascade are currently being targetted to reduce muscle wasting accompanying both acute metabolic derangements and chronic disorders such as cancer, congestive heart failure, sarcopenia, acquired immunodeficiency and long-lasting and poorly compensated diabetes mellitus.10
In all of these catabolic conditions, protein breakdown is enhanced and exceeds protein synthesis, resulting in loss of muscle mass and myofiber atrophy. There is substantial evidence indicating that autophagic flux is increased during muscle atrophy due to upregulation or downregulation of different sets of genes designated atrogenes.11, 12 These include genes encoding proteins that are degraded when autophagosomes fuse with lysosomes, such as the microtubule-associated protein 1 light chain 3 (LC3I). The latter is converted to LC3II during autophagy, so that the LC3II to LC3I ratio is often taken as representative of autophagy induction. However, whether autophagy itself causes muscle fiber degeneration remains a debated issue.13
Although TGF-beta1 has recently been shown to represent an important regulator of autophagy in liver and mammary cells,14 autophagy in the skeletal muscle is peculiar as compared with that occurring in other metabolically relevant tissues. During fasting, most tissues show a transient activation of autophagy. In contrast, skeletal muscle shows a persistent generation of autophagosomes that continues for days,11 suggesting that different mechanisms control autophagosome formation during short or long periods of induced autophagy. Whether and how TGF-beta1 features autophagy-promoting activity in muscle cells and whether this activity may lead to impaired myogenesis is unknown.
Phosphoprotein enriched in diabetes/phosphoprotein enriched in astrocytes (PED/PEA-15) is a 15 KDa molecule, which binds to different signaling proteins thereby regulating their function and several survival and metabolic transduction pathways.15, 16 PED/PEA-15 cellular levels are controlled by both transcriptional and post-translational mechanisms,16 which have been shown to be dysregulated in human disorders including type 2 diabetes. Many type 2 diabetic individuals feature exagerated expression of PED/PEA-15,17, 18 leading to impaired insulin action and secretion and, possibly, contributing to diabetes long-term complications.19 Quite recently, evidence has been reported indicating that PED/PEA-15 promotes autophagy in glioma cells through a JNK-dependent mechanism,20 rendering the cells resistant to starvation and other adverse stimuli. In the present work, we show that PED/PEA-15 autophagy-promoting action is necessary for enabling complete TGF-beta1 control of myogenesis.
Results
Effect of PED/PEA-15 on L6 cell differentiation
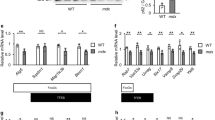
Low serum exposure causes L6 and C2C12 skeletal muscle cells to spontaneously differentiate in culture with the generation of multinucleate myotubes.7, 8, 9, 21 As previously reported,7, 8, 9 the L6 and C2C12 myotube formation is accompanied by upregulation of a number of genes including MyoD1, myogenin and GLUT4. Unexpectedly, however, these same gene profile studies revealed that expression of the autophagy regulator PED/PEA-15 underwent progressive reduction both during L6 and C2C12 cell differentiation (Figure 1a, left panel). Similar changes occurred in the case of PED/PEA-15 protein levels (right panel), raising the possibility that this multifunctional molecule exerts a previously unidentified physiological role in myogenesis. To test this hypothesis further, we have transfected L6 or C2C12 myoblasts with a PED/PEA-15 cDNA, increasing PED/PEA-15 levels by approximately 10-fold (Figure 1b, top panel). We then compared differentiation in these cells and in cells transfected with the empty vector. As shown in Figures 1b and c, PED/PEA-15 overexpression impaired myotube formation both in L6 and C2C12 and reduced activation of the myogenesis markers MyoD1, myogenin and GLUT4 by >2-fold in these cell types. At variance, the abundance of GLUT1 mRNA was 3-fold higher in PED/PEA-15-transfected as compared with the control cells.
Effect of PED/PEA-15 on L6 cell and C2C12 differentiation. (a) L6 and C2C12 cells were plated and differentiation was induced as described under Materials and Methods. Upon the indicated times in culture, the cells were lysed and protein samples were analysed by western blotting using PED/PEA-15 antibodies and beta-actin for control. Blots were revealed by ECL and autoradiography. Alternatively (left panel), total RNA was obtained from the cells and PED/PEA-15 mRNA was assessed by real-time RT-PCR using beta-actin as internal control. Each bar represents the mean ±S.D. of four independent experiments, each of which was performed in triplicate. (b) In L6 and C2C12 cells transfected with a cDNA encoding PED/PEA-15 (pcDNAIII-PED) or the empty vector (pcDNAIII), differentiation was induced as described under Materials and Methods. (c) Differentiated myotubes were then lysed and lysates blotted with PED/PEA-15 antibodies (top panel). Alternatively, total RNA was obtained from the cells and MyoD1, Myogenin, GLUT4, Glut1 and TGF-beta1 mRNAs were quantitated by real-time RT-PCR (bottom panel). Values are expressed as % of those in cells transfected with the empty vector. Each bar represents the mean ±S.D. of four independent experiments in triplicate. Asterisks denote statistically significant differences (*P<0.05; **P<0.01). Brightfield images of the L6 and C2C12 cells (100 × magnification) transfected with either the PED/PEA-15 cDNA or with the empty vector upon 6 days in culture. The microphotographs shown are representative of five other independent sets of observations
Role of PED/PEA-15 in TGF-beta1 action
To explore the role of PED/PEA-15 in muscle differentiation in greater detail, we have further examined its expression in L6 and C2C12 myoblasts upon addition of TGF-beta1 into their culture medium. Indeed, TGF-beta1 exerts a physiological role in controlling muscle development and constrains myogenesis in different cultured skeletal muscle cells.7, 8, 9 Interestingly, TGF-beta1 exposure enhanced PED/PEA-15 expression by almost 5-fold upon 6 h exposure (Figure 2a) and exhibited a consistent time- and dose-dependent effect on PED/PEA-15 protein levels as well (data not shown). The effect of TGF-beta1 on PED/PEA-15 expression was significantly reduced in L6 (or C2C12, data not shown) cells by the SB431542 inhibitor of the TGF-beta1 receptor, in parallel with SMAD3 phosphorylation (Figures 2b and c). Significant inhibition of TGF-beta1 effect was also achieved using a specific phosphorothioate antisense of SMAD3 (SMAD3 AS), indicating that TGF-beta1 stimulates PED/PEA-15 expression in a TGF-beta1R/SMAD3-dependent manner.
Effect of TGF-beta 1 on PED/PEA-15 expression in L6 and C2C12 cells. (a) Skeletal muscle cells were stimulated with 5 ng/ml TGF-beta1 for the indicated times. Total RNA was then isolated from the cells and the levels of PED/PEA-15 mRNA were assessed by real-time RT-PCR using beta-actin as internal control. Alternatively, the cells were treated with 0.1 μM of the TGF-beta1 receptor inhibitor SB431542 or transiently transfected with 200 nM SMAD3 phosphorothioate antisense (SMAD3 AS) followed by stimulation with 5 ng/ml TGF-beta1, as described under Materials and Methods. The cells were then harvested for assessment of PED/PEA-15 mRNA levels (b) or western blotted with specific PED/PEA-15, phospho-SMAD3 (pSMAD3) and SMAD3 antibodies (c). Blots were revealed by ECL and autoradiography using beta-actin as a loading control. The autoradiograps shown are representative of four independent experiments. Bars represent the mean ±S.D. of four independent experiments. Asterisks denote statistically significant differences (*P<0.05; **P<0.01; ***P<0.001)
On the basis of MyoD1, Myogenin and GLUT4 mRNA levels, TGF-beta1 treatment impaired L6 myoblasts differentiation (Figures 3a–c). This effect was largely prevented by two specific ShRNAs (ShPED1 and ShPED2), which silence PED/PEA-15 expression by >70% (Figures 3a–d). The effect of TGF-beta1 on myotube generation was similarly affected by the PED/PEA-15 ShRNA (Supplementary Figure 1). Phosphorylation of PED/PEA-15 was also required for TGF-beta1-mediated myoblasts differentiation. In fact, in presence of the PEDS116A mutant, TGF-beta1 failed to reduce MyoD1, Myogenin and GLUT4 mRNA levels (Figures 4a–d). Thus, in the L6 skeletal muscle cells, the complete regulatory role of TGF-beta1 on myogenesis requires the impact of TGF-beta1 signaling on PED/PEA-15. Same results were obtained in C2C12 (data not shown).
The role of PED/PEA-15 in TGF-beta1-impaired myogenesis in L6 cells. L6 myoblasts were transfected either with a scrambled or a PED/PEA-15 ShRNAs and maintained in culture enabling them to differentiate for the indicated times and in the presence or the absence of 5 ng/ml TGF-beta1. Total RNA preparations were then obtained from the cells and the levels of MyoD1 (a), Myogenin (b), Glut4 (c) and PED/PEA-15 mRNA (d) were quantitated by real-time RT-PCR analysis, using beta-actin as internal standard. Each bar represents the mean ±S.D. of four independent experiments in which each reaction was performed in triplicate. Asterisks denote statistically significant differences (#P<0.01 for differences between Sc at day 6 or 9 and Sc at day 0; *P<0.05; **P<0.01; ***P<0.001)
The role of PED/PEA-15 phosphorylation in TGF-beta1-impaired myogenesis in L6 cells. L6 cells transfected with the empty (pcDNAIII) or the PED/PEA-15 (pcDNAIII-PED) vector or the PED/PEA-15 S116A mutant (PEDS116A) were maintained in culture enabling them to differentiate for the indicated times and in the presence or the absence of 5 ng/ml TGF-beta1. Total RNA preparations were then obtained from the cells and the levels of MyoD1 (a), Myogenin (b), Glut4 (c) and PED/PEA-15 mRNA (d) were quantitated by real-time RT-PCR analysis, using beta-actin as internal standard. Each bar represents the mean ±S.D. of four independent experiments in which each reaction was performed in triplicate. Asterisks denote statistically significant differences (#P<0.01 for differences between Sc at day 6 and Sc at day 0; **P<0.01; ***P<0.001)
Promotion of autophagy by TGF-beta1 in L6 cells
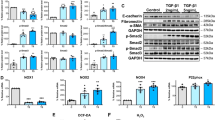
Current evidence indicates that fine tuning of autophagy has a major role in enabling differentiation.11, 12, 13 This observation led us to hypothesize that the recently recognized autophagy-promoting function of PED/PEA-1520 is necessary for the myogenesis constrain imposed by TGF-beta1, as TGF-beta1 has also been very recently shown to induce autophagy in non-muscle cells.14 FACS analysis of acridine orange-stained cells revealed that differentiating L6 (or C2C12, data not shown) cells treated with TGF-beta1 for 6 days accumulated acidic vescicular organelles (AVOs, a prominent feature of autophagy) to a significantly greater extent than control cells (Figure 5a). In parallel, TGF-beta1 treatment significantly increased both LC3I to LC3II conversion and Beclin 1 cell abundance, two known markers of autophagy activation (Figure 5b). In addition, exposure of L6 or C2C12 differentiating cells to the autophagy inhibitor 3-methyladenine led to rescue of the myotube differentiation markers in the presence of TGF-beta1 (Figure 5c), indicating that promotion of autophagy by TGF-beta1 is relevant to the differentiation constrain. Previous studies revealed that the forkhead transcription factor FoxO1 regulates the expression of several autophagy-related genes, including Beclin1 and LC3 and induces muscle wasting.22, 23, 24, 25 Consistently, exposure of L6 or C2C12 differentiating cells to TGF-beta1 led to dephosphorylation of both FoxO1 and of its upstream regulator, the PP2A serine/threonine phosphatase (Figure 5d).
TGF-beta1 effect on autophagy in L6 and C2C12 cells. (a) Differentiating L6 cells were treated with 5 ng/ml TGF-beta1 as described under Materials and methods. After 6 days in culture, the cells were stained with acridine orange and AVO accumulation was determined by flow cytometric analysis. Calculated percentages represent the red-positive cells in the upper quadrants (R10 and R11). FITC Lin, green color intensity; PE Texas Red Area, red color intensity. Those shown are representative of four independent experiments. Alternatively (b and d), L6 and C2C12 cells were lysed as described under Materials and Methods and western blotted using PED/PEA-15, Beclin1, LC3 I, LC3 II, phospho-Tyr307-PP2A, phospho-Ser256-FoxO1 or FoxO1 antibodies. Blots were revealed by ECL and autoradiographs, using beta-actin as loading control. The autoradiographs shown are representative of three independent experiments. (c) L6 and C2C12 cells were plated and exposed for 72 h to 5 ng/ml TGF-beta1 in the absence or the presence of 1 mM 3-methyladenine (3MA). Upon 6 days of differentiation, total RNA was isolated and the levels of MyoD1, Myogenin and Glut4 mRNA were quantitated by real-time RT-PCR analysis, with each reaction performed in triplicate. Bars represent the mean ±S.D. of four independent experiments. Asterisks denote statistically significant differences (*P<0.05; **P<0.01; ***P<0.001)
PED/PEA-15 promotion of autophagy mediates TGF-beta1 effect on differentiation in L6 cells
The overexpression of PED/PEA-15 (10-fold above the endogenous levels) was sufficient to determine a 10-fold increase in AVO accumulation, enhanced LC3I to LC3II conversion and augmentation in Beclin1 abundance (Figures 6a and b). As it is the case for myogenesis, ShPED1 (or ShPED2, data not shown) vector silencing of endogenous PED/PEA-15 impaired TGF-beta1 effects on both AVO accumulation and the autophagy markers (Figures 6c and d). Moreover, in L6 myoblasts, expression of exogenous PED/PEA-15 reduced PP2A and FoxO1 phosphorylation, whereas PED/PEA-15 silencing significantly attenuated TGF-beta1 effect on their phosphorylation (Figures 6b and d). At variance, transfection of the PEDS116A mutant did not elicit any effect on TGF-beta1-mediated autophagy as well as FoxO1 and PP2A dephosphorylation (data not shown).
PED/PEA-15 autophagy-promoting activity mediates TGF-beta1 action on L6 cell differentiation. L6 cells stably overexpressing PED/PEA-15 (pcDNAIII-PED) or transfected with the empty vector (pcDNAIII) were plated and enabled to differentiate as described under Materials and Methods. AVO accumulation was determined as outlined in Figure 4 (a). The one shown is representative of four independent experiments. (b) Alternatively, the cells were lysed and 100 microgram protein samples were analyzed by western blotting using the indicated antibodies. Blots were revealed by ECL and autoradiography, using beta-actin as loading control. The autoradiograph shown is representative of three independent experiments. (c) Differentiated myotubes were transiently transfected with either the ShPED/PEA-15 RNA (Sh PED/PEA-15) or a scrambled RNA (Sc) and then exposed to 5 ng/ml TGF-beta1. AVO accumulation was assessed as outlined in Figure 4. The one shown is representative of three independent experiments. (d) Alternatively, the cells were lysed and protein samples were analyzed by western blotting using the indicated antibodies. Blots were revealed by ECL and autoradiography, using beta-actin as loading control. The autoradiograph shown is representative of four independent experiments
Importantly, 3-methyladenine treatment of L6 differentiating cells stably overexpressing PED/PEA-15 reduced autophagy to levels similar to those detected in the untransfected cells and rescued myotube biochemical and morphological differentiation (Figures 7a–c). Same results were obtained in C2C12 (data not shown). Thus, the autophagy-promoting function of PED/PEA-15 appeared to mediate the induction of autophagy and the impaired myogenesis, which follows TGF-beta1 exposure.
3-methyladenine rescues differentiation in PED/PEA-15-overexpressing L6 cells. L6 myoblasts stably overexpressing PED/PEA-15 were plated and differentiated in culture as described under Materials and Methods. Seventy two hours before the experiment, the cells were exposed to 1 mM 3-methyladenine (3MA). AVO accumulation was then assessed as outlined under Figure 4 (a). The figures shown are representative of four independent experiments. Alternatively, total RNA was obtained by the cells followed by quantitation of MyoD1, Myogenin, GLUT4 and GLUT1 mRNA levels by RT-PCR, using beta-actin as internal standard (b). Data are presented as % of the levels measured in control cells. Bars represent the mean ±S.D. of four independent experiments in which each reaction was performed in triplicate. Asterisks denote statistically significant differences (**P<0.01; ***P<0.001). (c) Brightfield images (100 × ) of L6 cells stably transfected with either the PcDNAIII-PED vector or the empty PcDNAIII vector in the absence or the presence of 3-methyadenine. The microphotographs shown are representative of three additional experiments
In vivo consequences of skeletal muscle overexpression of PED/PEA-15
To address the significance of these findings to skeletal muscle in vivo, we performed histological analysis of tibialis anterior and quadriceps muscles from PED/PEA-15 transgenic mice. These animals have been previously described19, 26 and feature about 10-fold overexpression of the PED/PEA-15 gene in their muscles. As shown in Figures 8a–c, hematoxylin–eosin- and Engel trichrome-stained sections revealed a 10% increase in atrophic fibers between PED/PEA-15 transgenic mice and WT (panel a). Rimmed vacuoles (panel b) and vacuoles with red inclusions (panel c) were also significantly increased in the PED/PEA-15 transgenic muscles. Staining with specific antibodies revealed significantly higher expression of LC3 and Beclin1 in muscles form transgenic compared with those of control mice (Figures 8d and e). Indeed, muscles from the PED/PEA-15 transgenic mice revealed a >4-fold increase in the LC3II to LC3I ratio with similar-sized reduction in phospho-PP2A and phospho-FoxO1 levels (Figure 9a). MyoD1 and myogenin mRNA levels were also significantly reduced in muscles from the PED/PEA-15 transgenics (Figure 9b). Whether these biochemical changes were paralleled by in vivo functional abnormalities was further analyzed by determining transgenic mouse traveled distance in an open field. Interestingly, this test revealed a significant decrease in locomotor activity in the PED/PEA-15 transgenic mice (Figure 9c), suggesting a causal relationship between the muscle phenotype of these transgenic mice and the control of muscle development by PED/PEA-15.
Histological and immunohistochemical analysis of skeletal muscles from PED/PEA-15 transgenic mice. Sections from skeletal muscles (tibialis anterior) from 6-month-old transgenic mice and non-transgenic littermates (control; WT) were stained with either hematoxylin-eosin (H/E; a and b) or engel-trichrome (c) or immunostained with LC3 or Beclin1 (BECN1) antibodies (d, e). The arrows in the microphotographs (40 × ) indicate atrophic muscle fibers with basophilic cytoplasm (a), small rimmed vacuoles (b), vacuoles with red inclusions (c), decoration with LC3 (d) and Beclin1 (BECN1; e) antibodies. The microphotographs shown are representative of findings observations in six transgenic and eight non-transgenic animals
Functional consequences of PED/PEA-15 overexpression in skeletal muscle of transgenic mice. (a) Tibialis anterior muscles from PED/PEA-15 transgenic mice and from their non-transgenic littermates (WT) were dissected, solubilized and 100 μg protein samples were analyzed by western blotting using LC3-I, LC3-II, phospho-tyr307-PP2A (P-PP2A), PP2A, phospho-ser256-FoxO1 (p-FoxO1) and FoxO1 antibodies. Beta actin was used as loading control. Blots were revealed by ECL and autoradiography. The autoradiograph shown is representative of studies performed with n=8 mice/group. Alternatively (b), the muscle was used for obtaining total RNA preparation. MyoD1 and Myogenin mRNA was subsequently assessed by real-time RT-PCR analysis, using beta-actin as internal standard. Bars represent the mean±S.D. of four independent experiments each performed in triplicate. (c) Travel distance was compared in transgenic and non-transgenic littermates (16 animals/group) enabled to explore the open-field appartus for 5 min as described under Materials and methods. Bars represent the mean±S.D. of four independent studies each involving four mice/group. Asterisks denote statistically significant differences (*P<0.05; ***P<0.001)
Discussion
Skeletal muscle ontogeny and postnatal physiology are regulated by the TGF-beta family of cytokines in different animal systems from mice to humans. Significant advancement has also been made in the elucidation of the detailed mechanisms involved in this regulation.1, 2, 3, 4, 5, 6 However, many aspects of the TGF-beta signaling events leading to its action on skeletal muscle mass remain unsettled. In the present work, we have identified the multifunctional protein PED/PEA-15 as a key molecule transducing TGF-beta 1 signals and enabling TGF-beta1 repression of the myogenesis program.
We now report that, in the L6 and in C2C12 skeletal muscle cells the expression of PED/PEA-15 gene progressively declines during myogenesis. In addition, exogenous PED/PEA-15 overexpression inhibits myotubes generation and differentiation leading us to hypothesize that PED/PEA-15 is implicated in mechanisms controlling myogenesis. Indeed, we have further found that exposure to TGF-beta 1 restrains differentiation and, simultaneously, enhances PED/PEA-15 gene transcription significantly increasing abundance of PED/PEA-15 protein into the cell. This effect appears to be achieved via the TGF-beta1R/SMAD3 pathway activation, as inhibition or silencing of these key molecules significantly impaired PED/PEA-15 upregulation by TGF-beta1. Similar effects have been noted also in primary myoblasts from WT and TgPED mice (data not shown). Different Smad response elements were identified in the PED/PEA-15 promoter region whose occupancy increased following L6 cell exposure to TGF-beta1 (data not shown), suggesting regulation of PED/PEA-15 gene by TGF-beta1 signaling. Importantly, we show that ShRNAs silencing of endogenous PED/PEA-15 and abrogation of PED/PEA-15 phosphorylation at seryl 116 largely prevented TGF-beta1 action on muscle cell differentiation, indicating that PED/PEA-15 exerts a major role in mediating the myogenesis control exerted by TGF-beta1. The (TGF-beta1-mediated) phosphorylation status of PED/PEA-15 also may have a central role in this process because non-phosphorylated PED/PEA-15 lacks the capability to induce myogenic gene expression.
Despite intensive investigation, how TGF-beta1 inhibits myogenesis has been only partially elucidated. Smad3 is the key mediator,27, 28 as it physically interacts with both MyoD and MEF2C.29 The presence of PED/PEA-15 in this network was unanticipated and led us to explore its significance to TGF-beta1 action in greater detail. Autophagy is a dynamic process in which intracellular membrane structures, the autophagosomes, sequester proteins and organelles for degradation.30 Autophagy is implicated in homeostasis and cell-size regulation in multiple organs both during normal development and under stress conditions.31, 32 Recently, the role of autophagy in muscle differentiation has attracted much attention.11, 12, 13 However, it remains unclear whether autophagy is detrimental and part of the mechanisms that induce muscle degeneration, or whether it is a compensatory mechanism for cell survival. Both TGF-beta1 and PED/PEA-15 have been reported to activate autophagy in several tissues, though not in skeletal muscle.14, 20 In the present report, we describe that both of these molecules exert an autophagy-promoting action in the L6 skeletal muscle cells. Importantly, we obtained evidence that PED/PEA-15 promotion of autophagy mediates the effect of TGF-beta1 on differentiation in the L6 cells. Indeed, although silencing of the endogenous PED/PEA-15 blocked TGF-beta1-induced autophagy, inhibition with 3-methyladenine in L6 cells overexpressing PED/PEA-15 reduced autophagy to levels comparable to the untransfected cells and rescued myogenesis. All together, these findings led us to conclude that the autophagy-promoting function of PED/PEA-15 mediates both the autophagy induction and the impaired myogenesis caused by TGF-beta1. These effects have been obtained also in primary myoblasts from WT and TgPED mice (data not shown).
Very recent evidence indicates a direct role of the forkhead transcription factors (FoxOs) in regulating the cellular mechanisms responsible for autophagy activation and impairment of myoblasts differentiation. Foxo1 ablation in C2C12 cells or in mice increases the levels of MyoD, whereas forced expression of FoxO1 upregulates autophagic and atrophic responses in muscle cells.12, 22, 23, 24, 25, 33 Indeed, FoxOs have been shown to promote the expression of different autophagy-related genes. These include BECLIN1, ATG8, Gabarapl1, ATG12, ATG4B and VSP34.21, 22, 23, 24 FoxO1, in turn, is highly regulated at the post-transcriptional level. Dephosphorylation at Ser256 by the Ser/Thr phosphatase PP2A activates FoxO1, inducing its nuclear translocation.34, 35 Interestingly, PP2A has been shown to undergo activation in response to TGF-beta1 in different cell types.36 Here, we show that, in the L6 cells, full activation of the PP2A/FoxO1 signaling by TGF-beta1 requires PED/PEA-15. Indeed, (i) forcing PED/PEA-15 expression is sufficient to dephosphorylate PP2A and FoxO1, thus driving the nuclear translocation of FoxO1; and, (ii) PED/PEA-15 silencing significantly attenuates the effect of TGF-beta1 on the PP2A/FoxO1-transduced signal.34, 35, 36 How PED/PEA-15 impacts on PP2A has not been completely demonstrated yet. However, data from our own and other laboratories indicate that PED/PEA-15 induces PKC-delta in different cell types, including the L6 cells. PKC-delta, in turn, enhances PP2A activity,37, 38 indicating that it might induce PP2A in the L6 cell, as well.
These findings may have in vivo relevance as PP2A and FoxO1 activation accompanied by higher levels of autophagy markers and AVOs were also detected in skeletal muscle tissue from transgenic mice overexpressing PED/PEA-15. These changes were paralleled by detectable muscle atrophy and decreased locomotor activity in these animals. Indeed, there is now evidence that the FoxO family of transcription factors has a major transcriptional role in the muscle atrophy accompanying a number of disorders, including type 2 diabetes. In the type 2 diabetic individual, resistance to insulin in the PI3K/AKT signaling cascade is believed to represent a major mechanism responsible for muscle atrophy and sarcopenia, as impaired AKT function releases the brake on FoXO1 activity.34, 35 However, increased TGF-beta1 levels have also been reported in type 2 diabetes.19 Here, we show that TGF-beta1 upregulates PED/PEA-15 and may contribute to the increased PED/PEA-15 expression described in many individuals with type 2 diabetes.17, 18 This increased expression has no effect on PI3K/AKT signaling. Thus, TGF-beta1 induction of PED/PEA-15, may represent a previously unrecognized mechanism leading to FoxO1 activation, impaired myoblasts differentiation, muscle atrophy and decreased muscle mass, which accompany long-lasting diabetes. This hypothesis is currently under active investigation in our laboratory.
In conclusion, in the present report, we have identified PED/PEA-15 as novel molecule in the TGF-beta1 signaling network of the muscle cell. In this cell types, transcriptional upregulation of the PED/PEA-15 gene by TGF-beta1 induces autophagic responses, atrophy and might contribute to the decreased muscle mass accompanying long-lasting type 2 diabetes.
Materials and Methods
Materials
Media, sera, antibiotics for cell culture and the lipofectamine reagent were from Invitrogen (Grand Island, NY, USA). The PED/PEA-15 polyclonal antibody and pcDNAII-PED/PEA-15 vector have been previously described.19 The actin, pSMAD3, SMAD3, PP2A, pPP2A and Beclin1 antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA); the FoxO1 and pFoxO1 antibodies were from Upstate Biotechnolgy (Lake Placid, NY, USA); LC3 antibodies were from Cell Signaling Technology, Inc. (Beverly, MA, USA). TGF-beta1 cytokine was from PeproTech, Inc. (Rocky Hill, NJ, USA). The phosphorothioate oligonucleotides antisense sequences were as follows: SMAD3 AS: 5′-GCAGGATGGACGACAT-3′; SMAD3 S: 5′-GGAGTCAGACTGACGA-3′. Protein electrophoresis reagents were purchased from Bio-Rad (Richmond, VA, USA), western blotting and ECL reagents from Amersham Biosciences (Arlington Heights, IL, USA). PED/PEA-15 ShRNA vectors and all other chemicals were from Sigma (St. Louis, MO, USA).
Cell culture procedures and transfection
The L6 muscle and C2C12 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 10 000 μ/ml penicillin, 10 mg/ml streptomycin, and 2% l-glutamine in a humidified CO2 incubator and differentiated as described in Caruso et al.39 Stable transfection of the PED/PEA-15 cDNA and transient transfection of antisense oligonucleotides or PED/PEA-15 S116A mutant (PEDS116A) were performed by the lipofectamine method according to the manufacturer's instructions (Invitrogen). For these studies, 70 to 80% confluent cells were washed twice with Optimem (Invitrogen) and incubated for 8 h with 5 μg of plasmid construct, ShRNA vectors or antisense oligonucleotides (200 nM) and 45 to 60 μl of lipofectamine reagent. The medium was then replaced with DMEM with 10% (vol/vol) fetal bovine serum and cells further incubated for 15 h before being assayed. In transient transfection studies, the medium was then replaced with DMEM with 10% fetal bovine serum and cells were further incubated for 15 h before being assayed. For experiments with stably transfected cells, individual G418-resistant clones were selected by the limiting dilution technique (G418 effective dose, 0.8 mg/ml). The expression of PED/PEA-15 by the individual clones was quantitated by western blotting. In all transfection studies, the efficiency was consistently between 65 and 75%. Transfection efficiency of antisense oligonucleotides was estimated as 45±10% by co-transfection with green fluorescent protein.
TGF-beta1 treatment in differentiating L6 and C2C12 cells
When cells reached ∼90% of confluence (day 0), the medium was replaced with the differentiation-promoting medium (DMEM 2% fetal bovine serum), and the cells stimulated with TGF-beta1 (5 ng/ml) for 6 or 9 days (cytokine added on days 1, 3, 5 and 1, 3, 5, 7). Control cells were maintained in differentiation medium. The cells were then harvested and lysates were analyzed by western blotting, real time PCR, FACS analysis on days 6 and 9.
Mouse generation and phenotyping
The transgenic mice overexpressing PED/PEA-15 have been previously described.25 These mice were fertile. Their body weight is comparable to that of their wild-type littermates.25 All procedures described below were approved by the Institutional Animal Care and Utilisation Committee. Animals were kept in a 12-h dark–light cycle and had free access to standard diet. Mice chosen for experimentation were randomly selected from each box of mice housed in groups of three to four.
Open-field exploration
Free exploration in a square arena (walls made of transparent plexiglas 50 × 50 cm, 40 cm high) was allowed as previously described.40 Mice were enabled to explore for 5 min the open-field apparatus, placed in an experimental room with several large-scale environmental visual cues and homogeneous illumination. Exploratory behavior was recorded with a high definition digital camera and on-line videotracking was implemented using custom software written in Matlab environment. Total traveled distance was taken as an index of locomotor activity.
Muscle dissection and immunohistochemical examination
Skeletal muscle specimens (tibialis anterior or quadriceps) from PED/PEA-15 transgenic or control mice were processed according to Paciello et al.41 Briefly, dissected muscles were frozen in isopentane pre-cooled in liquid nitrogen and stored at 80 °C until being further analyzed. Sections (8 μm) were stained with haematoxylin and eosin (HE), Engel trichrome, or cytochrome C oxidase.
For immunohistochemical examination, frozen sections (6 μm) were fixed in acetone at 4 °C for 5 min, then blocked for endogenous peroxidase in 0.3% H2O2 in methanol for 20 min. Muscle sections were further incubated with mouse monoclonal antibodies against LC3 (dilution 1 : 50) and BECN1 (dilution 1 : 50) overnight at 4 °C. Slides were washed three times with PBS, incubated with biotinylated anti-rabbit and labeled streptavidin biotin (LSAB kit; DakoCytomation, Glostrup, Denmark) for 30 min, followed by incubation with streptavidin conjugated to horseradish peroxidase (LSAB Kit; DakoCytomation, Denmark). Color development was assessed following 5 min of diaminobenzidine treatment (DakoCytomation, Denmark) treatment. Sections were counterstained with Mayer's haematoxylin. In the corresponding negative control section, the primary antibody was either omitted or replaced with normal mouse serum.
Tissue and cell lysates and western blot analysis
Tissue samples were homogenised in a Polytron (Brinkman Instruments, Westbury, NY, USA) in 20 ml T-PER reagent (Pierce, Rockford, IL, USA) per gram of tissue according to the manufacturer's instructions. After centrifugation at 5000 × g for 5 min, the supernatant fractions were collected. Cells were solubilised in lysis buffer (50 mmol/l HEPES, pH 7.5, 150 mmol/l NaCl, 10 mmol/l EDTA, 10 mmol/l Na4P2O7, 2 mmol/l Na3VO4, 100 mmol/l NaF, 10% (vol/vol) glycerol, 1% (vol/vol) Triton X-100, 1 mmol/l PMSF, 10 mg/ml aprotinin) for 1 h at 4 °C and lysates were centrifuged at 5000 × g for 20 min. Total homogenates were separated by SDS-PAGE and transferred to 0.45 μm Immobilon-P membranes as previously described.20 Upon incubation with primary and secondary antibodies, immunoreactive bands were detected by ECL according to the manufacturer's instructions.
Real-time RT-PCR analysis
Total cellular RNA was isolated from muscles of wild-type and PED/PEA-15 transgenic mice using a commercial kit (RNeasy; Qiagen, Hilden, Germany) and according to the manufacturer's instructions. Tissue or cell RNA (1 μg) was reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen). PCR reactions were analysed using SYBR Green mix (Invitrogen). Reactions were performed using Platinum SYBR Green qPCR Super-UDG and a multicolour real-time PCR detection system (Cycler IQ; Bio-Rad). All reactions were performed in triplicate and β-actin was used as an internal standard. Primer sequences were as follows: TGF-beta1 (forward) 5′-ATACGCCTGAGTGGCTGTCT-3′, (reverse) 5′-TGGGACTGATCCCATTGATT-3′; β-actin (forward) 5′-GCGTGACATCAAAGAGAAG-3, (reverse) 5′-ACTGTGTTGGCATAGAGG-3′, MyoD1 (forward) 5′-TCGACTCACCAGACCTGCGCT-3′, (reverse) 5′-CTTGCAGGCCCACAGCAAGC; myogenin: (forward) 5′-ACTCCCTTACGTCCATCGTG-3′, (reverse) 5′-CAGGACAGCCCCACTTAAAA-3′, Glut4: (forward) 5′-CAGAAGGTGATTGAACAGAG-3′, (reverse) 5′-AATGATGCCAATGAGAAAGG-3′; Glut1: (forward) 5′-GGGAATGTCCTCATCTTGGA-3′, (reverse) 5′-TGAGGCTCTGTGTGGTTCTG-3′.
Quantification of acidic vesicular organelles with acridine orange
Autophagy in the L6 cells was assessed as previously described.42 Briefly, acidic vescicular organellles (AVOs) were quantified by acridine orange staining. For these studies cells were treated as necessary for each individual experiment and then gently detached with 0.05% trypsin–EDTA followed by 1.0 μg/ml acridine orange staining for 15 min at room temperature. Stained cells were then analyzed on a FACS cytometer (DakoCytomation, Carpinteria, CA, USA) and the Summit V4.3 software (DakoCytomation).42
Statistical methods
Data were analysed with Statview software (Abacus Concepts, Piscataway, NJ, USA) by one-factor analysis of variance. P-values of <0.05 were considered statistically significant.19
Abbreviations
- PED/PEA-15:
-
phosphoprotein enriched in diabetes/phosphoprotein enriched in astrocytes
- TGF-beta:
-
transforming growth factor-beta
- SMAD:
-
small mother against decapentaplegic
- PP2A:
-
protein phosphatase 2
- Foxo1:
-
forkhead box protein O1
- GLUT4:
-
glucose transporter type 4.
References
Ten Dijke P, Hill CS . New insights into TGF-beta-Smad signalling. Trends Biochem Sci 2004; 29: 265–273.
Massague J, Blain SW, Lo RS . TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 2000; 103: 295–309.
Dai C, Liu Y . Hepatocyte growth factor antagonizes the profibrotic action of TGF-β1 in mesangial cells by stabilizing Smad transcriptional corepressor TGIF. J Am Soc Nephrol 2004; 15: 1402–1412.
Letterio JJ, Roberts AB . Regulation of immune responses by TGF-beta. Annu Rev Immunol 1998; 16: 137–161.
Ha H, Yu MR, Lee HB . High glucose-induced PKC activation mediates TGF-beta 1 and fibronectin synthesis by peritoneal mesothelial cells. Kidney Int 2001; 59: 463–470.
Toda S, Matsumura S, Fujitani N, Nishimura T, Yonemitsu N, Sugihara H . Transforming growth factor-beta1 induces a mesenchyme-like cell shape without epithelial polarization in thyrocytes and inhibits thyroid folliculogenesis in collagen gel culture. Endocrinology 1997; 138: 5561–5575.
Massague J, Cheifetz S, Endo T, Nadal-Ginard B . Type beta transforming growth factor is an inhibitor of myogenic differentiation. Proc Natl Acad Sci 1986; 83: 8206–8210.
Kollias HD, McDermott JC . Transforming growth factor-beta and myostatin signaling in skeletal muscle. J Appl Physiol 2008; 104: 579–587.
Schabort EJ, Van Der Merwe M, Loos B, Moore FP, Niesler CU . TGF-beta's delay skeletal muscle progenitor cell differentiation in an isoform-independent manner. Exp Cell Res 2009; 315: 373–384.
Glass DJ . Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 2005; 37: 1974–1984.
Sandri M . Autophagy in skeletal muscle. FEBS 2010; 584: 1411–1416.
Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007; 6: 472–483.
Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M et al. Autophagy is required to maintain muscle mass. Cell Metab 2009; 10: 507–515.
Suzuki HI, Kiyono K, Miyazono K . Regulation of autophagy by transforming growth factor-beta (TGFbeta) signaling. Autophagy 2010; 14: 6.
Araujo H, Danziger N, Cordier J, Glowinski J, Chneiweiss H . Characterization of PEA-15 a major substrate for protein kinase C in astrocytes. J Biol Chem 1993; 268: 5911–5920.
Fiory F, Formisano P, Perruolo G, Beguinot F . Frontiers: PED/PEA-15, a multifunctional protein controlling cell survival and glucose metabolism. Am J Physiol Endocrinol Metab 2009; 297: 592–601.
Condorelli G, Vigliotta G, Iavarone C, Caruso M, Tocchetti CG, Andreozzi F et al. PED/PEA-15 gene controls glucose transport and is overexpressed in type 2 diabetes mellitus. EMBO J 1998; 7: 3858–3866.
Valentino R, Lupoli G, Raciti GA, Oriente F, Farinaro E, Della Valle E et al. The PEA15 gene is overexpressed and related to insulin resistance in healthy first degree relatives of patients with type 2 diabetes. Diabetologia 2006; 49: 3058–3066.
Oriente F, Iovino S, Cassese A, Romano C, Miele C, Troncone G et al. Overproduction of phosphoprotein enriched in diabetes (PED) induces mesangial expansion and upregulates protein kinase C-beta activity and TGF-beta1 expression. Diabetologia 2009; 52: 2642–2652.
Böck BC, Tagscherer KE, Fassl A, Krämer A, Oehme I, Zentgraf HW et al. The PEA-15 protein regulates autophagy via activation of JNK. J Biol Chem 2010; 285: 21644–21654.
Pisani DF, Cabane C, Derijard B, Dechesne CA . The topoisomerase 1-interacting protein BTBD1 is essential for muscle cell differentiation. Cell Death Differ 2004; 11: 1157–1165.
Salih DA, Brunet A . FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 2008; 20: 126–136.
Sengupta A, Molkentin JD, Yutzey KE . FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem 2009; 284: 28319–28331.
Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004; 117: 399–412.
Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 2004; 279: 41114–41123.
Vigliotta G, Miele C, Santopietro S, Portella G, Perfetti A, Maitan MA et al. Overexpression of the ped/pea-15 gene causes diabetes by impairing glucose-stimulated insulin secretion in addition to insulin action. Mol Cell Biol 2004; 24: 5005–5015.
Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ . Myostatin reduces Akt/TORC1/p70S6 K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol 2009; 296: 1258–1270.
Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R et al. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol 2009; 296: 1248–1257.
Kollias HD, McDermott JC . Transforming growth factor-b and myostatin signaling in skeletal muscle. J Appl Physiol 2007; 104: 579–587.
Meléndez A, Neufeld TP . The cell biology of autophagy in metazoans: a developing story. Development 2008; 135: 2347–2360.
Shintani T, Klionsky DJ . Autophagy in health and disease: a double-edged sword. Science 2004; 306: 990–995.
Vellai T, Bicsák B, Tóth ML, Takács-Vellai K, Kovács AL . Regulation of cell growth by autophagy. Autophagy 2008; 4: 507–509.
Liu CM, Yang Z, Liu CW, Wang R, Tien P, Dale R et al. Effect of RNA oligonucleotide targeting Foxo-1 on muscle growth in normal and cancer cachexia mice. Cancer Gene Ther 2007; 14: 945–952.
Yan L, Lavin VA, Moser LR, Cui Q, Kanies C, Yang E . PP2A regulates the pro-apoptotic activity of FOXO1. J Biol Chem 2008; 283: 7411–7420.
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96: 857–868.
Petritsch C, Beug H, Balmain A, Oft M . TGF-beta inhibits p70 S6 kinase via protein phosphatase 2A to induce G(1) arrest. Genes Dev 2000; 14: 3093–3101.
Renault-Mihara F, Beuvon F, Iturrioz X, Canton B, De Bouard S, Léonard N et al. Phosphoprotein enriched in astrocytes-15 kDa expression inhibits astrocyte migration by a protein kinase C delta-dependent mechanism. Mol Biol Cell 2006; 17: 5141–5152.
Zhang D, Kanthasamy A, Yang Y, Anantharam V, Kanthasamy A . Protein kinase C delta negatively regulates tyrosine hydroxylase activity and dopamine synthesis by enhancing protein phosphatase-2A activity in dopaminergic neurons. J Neurosci 2007; 27: 5349–5362.
Caruso M, Miele C, Formisano P, Condorelli G, Bifulco G, Oliva A et al. In skeletal muscle, glucose storage and oxidation are differentially impaired by the IR1152 mutant receptor. J Biol Chem 1997; 272: 7290–7297.
Mignogna P, Viggiano D . Brain distribution of genes related to changes in locomotor activity. Physiol Behav 2010; 19: 618–626.
Paciello O, Papparella S . Histochemical and immunohistological approach to comparative neuromuscular diseases. Folia Histochem Cytobiol 2009; 47: 143–152.
Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990; 82: 1107–1112.
Acknowledgements
This work was supported by the European Foundation for the Study of Diabetes (EFSD), the European Community's PREPOBEDIA (201681), grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC), and from the Ministero dell’Università e della Ricerca Scientifica (MERIT initiative, FIRB program: RBNE08NKH7). The financial contribution of Telethon Italy is gratefully acknowledged.
Author Contributions
SI and FO generated data, discussion, and wrote the manuscript. GB researched data, performed experimental work, and contributed to discussion. SC, VI, DV and GP, researched data, OP contributed to critical discussion and researched data, PF and FB directed and planned research and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by RA Knight
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Iovino, S., Oriente, F., Botta, G. et al. PED/PEA-15 induces autophagy and mediates TGF-beta1 effect on muscle cell differentiation. Cell Death Differ 19, 1127–1138 (2012). https://doi.org/10.1038/cdd.2011.201
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2011.201
Keywords
This article is cited by
-
Inhibition of the JNK/MAPK signaling pathway by myogenesis-associated miRNAs is required for skeletal muscle development
Cell Death & Differentiation (2018)
-
Parkinson-like phenotype in insulin-resistant PED/PEA-15 transgenic mice
Scientific Reports (2016)
-
Inhibition of miR-155 Protects Against LPS-induced Cardiac Dysfunction and Apoptosis in Mice
Molecular Therapy - Nucleic Acids (2016)
-
Autophagy regulates odontoblast differentiation by suppressing NF-κB activation in an inflammatory environment
Cell Death & Disease (2016)
-
Enhancement of chondrocyte autophagy is an early response in the degenerative cartilage of the temporomandibular joint to biomechanical dental stimulation
Apoptosis (2013)