Abstract
Huntington's disease (HD) is an autosomal-dominant neurological disorder caused by expanded CAG repeats in the Huntingtin (Htt) gene, but it is not known how this mutation causes neurodegeneration. Herein, we found that dysfunction of upstream binding factor-1 (UBF-1) is linked to reduced ribosomal DNA (rDNA) transcription in HD. We identified that UBF1 acetylation at Lys (K) 352 by CREB binding protein (CBP) is crucial for the transcriptional activity of rDNA. UBF1 mutation (K352A, K352Q, and K352R) decreased rDNA transcriptional activity. Moreover, both CBP–dHAT mutant and knockdown of CBP by siRNA reduced acetylation of UBF1 and resulted in the decreased transcription of rDNA into rRNA. ChIP analysis showed a significant reduction of UBF1 occupancy in the promoter of rDNA in STHdhQ111 cell line model of HD. These results demonstrate that abnormal activity of UBF1 and its acetylation by CBP are linked to impaired rDNA transcription in HD. This novel mechanism suggests that modulation of UBF-mediated rDNA synthesis by CBP may be a therapeutic target for improving neuronal rDNA transcription in HD.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
Abbreviations
- ENH:
-
enhancer
- UCE:
-
upstream control element
- CORE:
-
core region
- ETS:
-
external transcribed spacer
- 5-FU:
-
5-fluorouracil
References
Vonsattel JP, DiFiglia M . Huntington disease. J Neuropathol Exp Neurol 1998; 57: 369–384.
Sadri-Vakili G, Cha JH . Mechanisms of disease: Histone modifications in Huntington's disease. Nat Clin Pract Neurol 2006; 2: 330–338.
Steffan JS, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 2001; 413: 739–743.
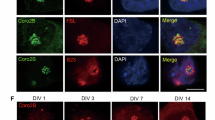
Casafont I, Bengoechea R, Navascués J, Pena E, Berciano MT, Lafarga M . The giant fibrillar center: a nucleolar structure enriched in upstream binding factor (UBF) that appears in transcriptionally more active sensory ganglia neurons. J Struct Biol 2007; 159: 451–461.
Lafontaine DL, Tollervey D . The function and synthesis of ribosomes. Nat Rev Mol Cell Biol 2001; 2: 514–520.
Akhmanova A, Verkerk T, Langeveld A, Grosveld F, Galjart N . Characterisation of transcriptionally active and inactive chromatin domains in neurons. J Cell Sci 2000; 113: 4463–4474.
McStay B, Grummt I . The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 2008; 24: 131–157.
Hirschler-Laszkiewicz I, Cavanaugh A, Hu Q, Catania J, Avantaggiati ML, Rothblum LI . The role of acetylation in rDNA transcription. Nucleic Acids Res 2001; 29: 4114–4124.
Yamada M, Sato T, Shimohata T, Hayashi S, Igarashi S, Tsuji S et al. Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases. Am J Pathol 2001; 159: 1785–1795.
Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C et al. Exon1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 1996; 87: 493–506.
Pelletier G, Stefanovsky VY, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum LI et al. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell 2000; 6: 1059–1066.
Lee J, Hagerty S, Cormier KA, Kim J, Kung AL, Ferrante RJ et al. Monoallele deletion of CBP leads to pericentromeric heterochromatin condensation through ESET expression and histone H3 (K9) methylation. Hum Mol Genet 2008; 17: 1774–1782.
Stefanovsky VY, Moss T . The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation. Nucleic Acids Res 2008; 36: 5093–5101.
O'Mahony DJ, Rothblum LI . Identification of two forms of the RNA polymerase I transcription factor UBF. Proc Natl Acad Sci USA 1991; 88: 3180–3184.
Hannan RD, Stefanovsky V, Taylor L, Moss T, Rothblum LI . Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocyte: implications for cardiac hypertrophy. Proc Natl Acad Sci USA 1996; 93: 8750–8755.
Chen D, Belmont AS, Huang S . Upstream binding factor association induces large-scale chromatin decondensation. Proc Natl Acad Sci USA 2004; 101: 15106–15111.
Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol 2008; 183: 1259–1274.
Meraner J, Lechner M, Loidl A, Goralik-Schramel M, Voit R, Grummt I et al. Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res 2006; 34: 1798–1806.
Lin CH, Platt MD, Ficarro SB, Hoofnagle MH, Shabanowitz J, Comai L et al. Mass spectrometric identification of phosphorylation sites of rRNA transcription factor upstream binding factor. Am J Physiol Cell Physiol 2007; 292: 1617–1624.
Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H et al. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA 2000; 97: 6763–6768.
Zoghbi HY, Orr HT . Glutamine repeats and neurodegeneration. Annu Rev Neurosci 2000; 23: 217–247.
McCampbell A, Fischbeck KH . Polyglutamine and CBP: fatal attraction? Nat Med 2001; 7: 528–530.
Nucifora FC, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M et al. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 2001; 291: 2423–2428.
Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G et al. Histone deacetylase 1 can repress transcription by binding to SP1. Mol Cell Biol 1999; 19: 5504–5511.
Stefanovsky VY, Moss T . Regulation of rRNA synthesis in human and mouse cells is not determined by changes in active gene count. Cell Cycle 2006; 5: 735–739.
Ryu H, Lee J, Olofsson BA, Mwidau A, Dedeoglu A, Escudero M et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci USA 2003; 100: 4281–4286.
McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH . Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci USA 2001; 98: 15179–15184.
Hughes RE, Olson JM . Therapeutic opportunities in polyglutamine disease. Nat Med 2001; 7: 419–423.
Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM et al. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 2004; 279: 6783–6793.
Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet 2000; 9: 2799–2809.
Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci USA 2006; 103: 19176–19181.
Ryu H, Lee J, Zaman K, Kubilis J, Ferrante RJ, Ross BD et al. Sp1 and Sp3 are oxidative stress-inducible, anti-death transcription factors in cortical neurons. J Neurosci 2003; 23: 3597–3606.
Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ . Antioxidants modulate mitochondrial protein kinase A and increase CREB binding to D-Loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci USA 2005; 102: 13915–13920.
Kim T, Kim SJ, Kim K, Kang UB, Lee C, Park KS et al. Profiling of vitreous proteomes from proliferative diabetic retinopathy and nondiabetic patients. Proteomics 2007; 7: 4203–4215.
Garaguso I, Borlak J . Matrix layer sample preparation: an improved MALDI-MS peptide analysis method for proteomic studies. Proteomics 2008; 8: 2583–2595.
Ryu H, Smith K, Camelo SI, Carreras I, Lee J, Iglesias AH et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem 2005; 93: 1087–1098.
Acknowledgements
We thank Dr. Samson T. Jacob for pIRES-Luc and Human rRNA-luciferase Vector and Dr. Marcy MacDonald for STHdhQ7/7 and STHdhQ111/111 cells. We thank Dr. Byoung Yul Soh for his technical assistance. This study was supported by WCU Neurocytomics Program Grant (800-20080848) (H.R.) and SRC Grant (2010-0029-403) (H.R.) from KOSEF and NIH NS52724 (H.R.). This work was also supported in part by the Converging Research Center Program (20090094081) (Y.K.) through NRF of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by N Bazan
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, J., Hwang, Y., Boo, J. et al. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington's disease. Cell Death Differ 18, 1726–1735 (2011). https://doi.org/10.1038/cdd.2011.38
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2011.38
Keywords
This article is cited by
-
PQBP5/NOL10 maintains and anchors the nucleolus under physiological and osmotic stress conditions
Nature Communications (2023)
-
Proapoptotic Requirement of Ribosomal Protein L11 in Ribosomal Stress-Challenged Cortical Neurons
Molecular Neurobiology (2018)
-
Dopamine perturbation of gene co-expression networks reveals differential response in schizophrenia for translational machinery
Translational Psychiatry (2018)
-
Transcription, Epigenetics and Ameliorative Strategies in Huntington’s Disease: a Genome-Wide Perspective
Molecular Neurobiology (2015)
-
Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease
Cell & Bioscience (2014)