Abstract
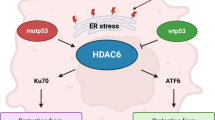
Mutant p53 (mutp53) cancers are surprisingly dependent on their hyperstable mutp53 protein for survival, identifying mutp53 as a potentially significant clinical target. However, exploration of effective small molecule therapies targeting mutp53 has barely begun. Mutp53 hyperstabilization, a hallmark of p53 mutation, is cancer cell-specific and due to massive upregulation of the HSP90 chaperone machinery during malignant transformation. We recently showed that stable complex formation between HSP90 and its mutp53 client inhibits E3 ligases MDM2 and CHIP, causing mutp53 stabilization. Histone deacetylase (HDAC) inhibitors (HDACi) are a new class of promising anti-cancer drugs, hyperacetylating histone and non-histone targets. Currently, suberoylanilide hydroxamic acid (SAHA) is the only FDA-approved HDACi. We show that SAHA exhibits preferential cytotoxicity for mutant, rather than wild-type and null p53 human cancer cells. Loss/gain-of-function experiments revealed that although able to exert multiple cellular effects, SAHA's cytotoxicity is caused to a significant degree by its ability to strongly destabilize mutp53 at the level of protein degradation. The underlying mechanism is SAHA's inhibition of HDAC6, an essential positive regulator of HSP90. This releases mutp53 and enables its MDM2- and CHIP-mediated degradation. SAHA also strongly chemosensitizes mutp53 cancer cells for chemotherapy due to its ability to degrade mutp53. This identifies a novel action of SAHA with the prospect of SAHA becoming a centerpiece in mutp53-specific anticancer strategies.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- wtp53:
-
wild-type p53
- mutp53:
-
mutant p53
- GOF:
-
gain-of-function
- HDAC:
-
histone deacetylase
- HDACi:
-
histone deacetylase inhibitor
- CHX:
-
cycloheximide
- KI:
-
knock-in
- SFN:
-
Sulforaphane
- SAHA:
-
suberoylanilide hydroxamic acid
References
Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004; 119: 861–872.
Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004; 119: 847–860.
Blandino G, Levine AJ, Oren M . Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 1999; 18: 477–485.
Li R, Sutphin PD, Schwartz D, Matas D, Almog N, Wolkowicz R et al. Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene 1998; 16: 3269–3277.
Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A . Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene 2006; 25: 304–309.
Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 2008; 22: 1337–1344.
Li D, Marchenko N, Schulz R, Fischer V, Velasco-Hernandez T, Talos F et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 2011; 9: 577–588.
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 2009; 137: 87–98.
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139: 1327–1341.
Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin Jr WG . Chemosensitivity linked to p73 function. Cancer Cell 2003; 3: 403–410.
Minucci S, Pelicci PG . Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 2006; 6: 38–51.
Bali P, George P, Cohen P, Tao J, Guo F, Sigua C et al. Superior activity of the combination of histone deacetylase inhibitor LAQ824 and the FLT-3 kinase inhibitor PKC412 against human acute myelogenous leukemia cells with mutant FLT-3. Clin Cancer Res 2004; 10: 4991–4997.
Fuino L, Bali P, Wittmann S, Donapaty S, Guo F, Yamaguchi H et al. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol Cancer Ther 2003; 2: 971–984.
Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J et al. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or -refractory chronic myelogenous leukemia-blast crisis cells. Cancer Res 2003; 63: 5126–5135.
Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA et al. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst 2002; 94: 504–513.
Bolden JE, Peart MJ, Johnstone RW . Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 2006; 5: 769–784.
Esser C, Scheffner M, Hohfeld J . The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem 2005; 280: 27443–27448.
Lukashchuk N, Vousden KH . Ubiquitination and degradation of mutant p53. Mol Cell Biol 2007; 27: 8284–8295.
Whitesell L, Sutphin P, An WG, Schulte T, Blagosklonny MV, Neckers L . Geldanamycin-stimulated destabilization of mutated p53 is mediated by the proteasome in vivo. Oncogene 1997; 14: 2809–2816.
Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 2005; 18: 601–607.
Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell 2007; 25: 151–159.
Gibbs A, Schwartzman J, Deng V, Alumkal J . Sulforaphane destabilizes the androgen receptor in prostate cancer cells by inactivating histone deacetylase 6. Proc Natl Acad Sci USA 2009; 106: 16663–16668.
Blagosklonny MV, Trostel S, Kayastha G, Demidenko ZN, Vassilev LT, Romanova LY et al. Depletion of mutant p53 and cytotoxicity of histone deacetylase inhibitors. Cancer Res 2005; 65: 7386–7392.
Gui CY, Ngo L, Xu WS, Richon VM, Marks PA . Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA 2004; 101: 1241–1246.
Varshochi R, Halim F, Sunters A, Alao JP, Madureira PA, Hart SM et al. ICI182,780 induces p21Waf1 gene transcription through releasing histone deacetylase 1 and estrogen receptor alpha from Sp1 sites to induce cell cycle arrest in MCF-7 breast cancer cell line. J Biol Chem 2005; 280: 3185–3196.
Whitesell L, Lindquist SL . HSP90 and the chaperoning of cancer. Nat Rev Cancer 2005; 5: 761–772.
Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH . The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol 1998; 18: 1517–1524.
Sayan BS, Yang AL, Conforti F, Bernardini S, Tucci P, Vasa-Nicotera M et al. Induction of Tap63 by histone deacetylase inhibitors. Biochem Biophys Res Commun 2010; 391: 1748–1751.
Beyer U, Moll-Rocek J, Moll UM, Dobbelstein M . Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc Natl Acad Sci USA 2011; 108: 3624–3629.
Yan W, Liu G, Scoumanne A, Chen X . Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer Res 2008; 68: 6789–6796.
Goh AM, Coffill CR, Lane DP . The role of mutant p53 in human cancer. J Pathol 2011; 223: 116–126.
King FW, Wawrzynow A, Hohfeld J, Zylicz M . Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J 2001; 20: 6297–6305.
Tsutsumi S, Neckers L . Extracellular heat shock protein 90: a role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci 2007; 98: 1536–1539.
Yi X, Wei W, Wang SY, Du ZY, Xu YJ, Yu XD . Histone deacetylase inhibitor SAHA induces ERalpha degradation in breast cancer MCF-7 cells by CHIP-mediated ubiquitin pathway and inhibits survival signaling. Biochem Pharmacol 2008; 75: 1697–1705.
Muhlenberg T, Zhang Y, Wagner AJ, Grabellus F, Bradner J, Taeger G et al. Inhibitors of deacetylases suppress oncogenic KIT signaling, acetylate HSP90, and induce apoptosis in gastrointestinal stromal tumors. Cancer Res 2009; 69: 6941–6950.
Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 2007; 110: 313–322.
Lindemann RK, Newbold A, Whitecross KF, Cluse LA, Frew AJ, Ellis L et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci USA 2007; 104: 8071–8076.
Acknowledgements
This work was funded by grants from the National Cancer Institute (CA0664 and CA93853 to UMM), Deutsche Krebshilfe (108173 to UMM), Deutsche Forschungsgemeinschaft (MO 1998/1-1 to UMM) and the Carol Baldwin Breast Cancer Research Fund (to NDM). We also thank Sulan Xu, Alisha Yalowitz and Chia-Ying Yang (Stony Brook University) for their technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Li, D., Marchenko, N. & Moll, U. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ 18, 1904–1913 (2011). https://doi.org/10.1038/cdd.2011.71
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2011.71
Keywords
This article is cited by
-
Translating p53-based therapies for cancer into the clinic
Nature Reviews Cancer (2024)
-
Precise pancreatic cancer therapy through targeted degradation of mutant p53 protein by cerium oxide nanoparticles
Journal of Nanobiotechnology (2023)
-
HDACs/mTOR inhibitor synergizes with pyrotinib in HER2-positive pancreatic cancer through degradation of mutant P53
Cancer Cell International (2022)
-
FK228 potentiates topotecan activity against small cell lung cancer cells via induction of SLFN11
Acta Pharmacologica Sinica (2022)
-
Should mutant TP53 be targeted for cancer therapy?
Cell Death & Differentiation (2022)