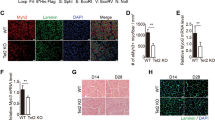
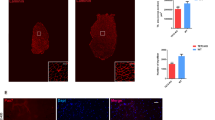
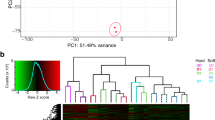
Abstract
The TEAD (1–4) transcription factors comprise the conserved TEA/ATTS DNA-binding domain recognising the MCAT element in the promoters of muscle-specific genes. Despite extensive genetic analysis, the function of TEAD factors in muscle differentiation has proved elusive due to redundancy among the family members. Expression of the TEA/ATTS DNA-binding domain that acts as a dominant negative repressor of TEAD factors in C2C12 myoblasts inhibits their differentiation, whereas selective shRNA knockdown of TEAD4 results in abnormal differentiation characterised by the formation of shortened myotubes. Chromatin immunoprecipitation coupled to array hybridisation shows that TEAD4 occupies 867 promoters including those of myogenic miRNAs. We show that TEAD factors directly induce Myogenin, CDKN1A and Caveolin 3 expression to promote myoblast differentiation. RNA-seq identifies a set of genes whose expression is strongly reduced upon TEAD4 knockdown among which are structural and regulatory proteins and those required for the unfolded protein response. In contrast, TEAD4 represses expression of the growth factor CTGF (connective tissue growth factor) to promote differentiation. Together these results show that TEAD factor activity is essential for normal C2C12 cell differentiation and suggest a role for TEAD4 in regulating expression of the unfolded protein response genes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- TEA:
-
Yeast (TEC-1), Aspergillus nidulans (AbaA) and Drosophilla (scalloped)
- ATTS:
-
Aspergillus nidulans (AbaA), Yeast (TEC-1), human TEF1, and Drosophilla (scalloped)
- DBD:
-
DNA-binding domain
- SV40:
-
simian virus 40
- CTGF:
-
connective tissue growth factor
- β-MHC, myosin, heavy polypeptide 7, cardiac muscle, beta; YAP1:
-
Yes-associated protein 1
- WWTR1:
-
WW domain containing transcription regulator 1
- ChIP-chip:
-
chromatin immunoprecipitation-array hybridization
- Flag:
-
FLAG octapeptide tag
- HA:
-
Hemagglutinin tag
- TSS:
-
transcription start site
- UPR:
-
unfolded protein response
- Cdkn1a:
-
cyclin-dependent kinase inhibitor 1A (P21)
- TRP53:
-
transformation-related protein 53
- SP1:
-
trans-acting transcription factor 1
- RT-qPCR:
-
reverse transcription–quantitative polymerase chain reaction
- VGL:
-
vestigial
References
Jacquemin P, Hwang JJ, Martial JA, Dolle P, Davidson I . A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem 1996; 271: 21775–21785.
Yoshida T . MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol 2008; 28: 8–17.
Burglin TR . The TEA domain: a novel, highly conserved DNA-binding motif [letter]. Cell 1991; 66: 11–12.
Andrianopoulos A, Timberlake WE . ATTS, a new and conserved DNA binding domain [letter]. Plant Cell 1991; 3: 747–748.
Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S . Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci USA 2006; 103: 17225–17230.
Mar JH, Ordahl CP . A conserved CATTCCT motif is required for skeletal muscle-specific activity of the cardiac troponin T gene promoter. Proc Natl Acad Sci USA 1988; 85: 6404–6408.
Chen Z, Friedrich GA, Soriano P . Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev 1994; 8: 2293–2301.
Chen HH, Baty CJ, Maeda T, Brooks S, Baker LC, Ueyama T et al. Transcription enhancer factor-1-related factor-transgenic mice develop cardiac conduction defects associated with altered connexin phosphorylation. Circulation 2004; 110: 2980–2987.
Maeda T, Mazzulli JR, Farrance IK, Stewart AF . Mouse DTEF-1 (ETFR-1, TEF-5) is a transcriptional activator in alpha 1-adrenergic agonist-stimulated cardiac myocytes. J Biol Chem 2002; 277: 24346–24352.
Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD . An initial blueprint for myogenic differentiation. Genes Dev 2005; 19: 553–569.
Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007; 134: 3827–3836.
Zhao B, Li L, Lei Q, Guan KL . The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 2010; 24: 862–874.
Hwang JJ, Chambon P, Davidson I . Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J 1993; 12: 2337–2348.
Montano MM, Lim RW . Glucocorticoid effects on the skeletal muscle differentiation program: analysis of clonal proliferation, morphological differentiation and the expression of muscle-specific and regulatory genes. Endocr Res 1997; 23: 37–57.
Anderson C, Catoe H, Werner R . MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 2006; 34: 5863–5871.
Guo K, Wang J, Andres V, Smith RC, Walsh K . MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol 1995; 15: 3823–3829.
Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ et al. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 1995; 267: 1018–1021.
Sugiura K, Muro Y, Futamura K, Matsumoto K, Hashimoto N, Nishizawa Y et al. The unfolded protein response is activated in differentiating epidermal keratinocytes. J Invest Dermatol 2009; 129: 2126–2135.
Nakanishi K, Sudo T, Morishima N . Endoplasmic reticulum stress signaling transmitted by ATF6 mediates apoptosis during muscle development. J Cell Biol 2005; 169: 555–560.
Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 2007; 27: 53–66.
Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K . XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001; 107: 881–891.
Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 2002; 16: 452–466.
Harding HP, Calfon M, Urano F, Novoa I, Ron D . Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol 2002; 18: 575–599.
Sambasivan R, Cheedipudi S, Pasupuleti N, Saleh A, Pavlath GK, Dhawan J . The small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoter. J Cell Sci 2009; 122: 3481–3491.
Vietor I, Vadivelu SK, Wick N, Hoffman R, Cotten M, Seiser C et al. TIS7 interacts with the mammalian SIN3 histone deacetylase complex in epithelial cells. Embo J 2002; 21: 4621–4631.
Vadivelu SK, Kurzbauer R, Dieplinger B, Zweyer M, Schafer R, Wernig A et al. Muscle regeneration and myogenic differentiation defects in mice lacking TIS7. Mol Cell Biol 2004; 24: 3514–3525.
Cao X, Pfaff SL, Gage FH . YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev 2008; 22: 3320–3334.
Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 2010; 18: 662–674.
Aziz A, Liu QC, Dilworth FJ . Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle. Epigenetics 2010; 5: 691–695.
Sok J, Wang XZ, Batchvarova N, Kuroda M, Harding H, Ron D . CHOP-Dependent stress-inducible expression of a novel form of carbonic anhydrase VI. Mol Cell Biol 1999; 19: 495–504.
Goruppi S, Patten RD, Force T, Kyriakis JM . Helix-loop-helix protein p8, a transcriptional regulator required for cardiomyocyte hypertrophy and cardiac fibroblast matrix metalloprotease induction. Mol Cell Biol 2007; 27: 993–1006.
Kong DK, Georgescu SP, Cano C, Aronovitz MJ, Iovanna JL, Patten RD et al. Deficiency of the transcriptional regulator p8 results in increased autophagy and apoptosis, and causes impaired heart function. Mol Biol Cell 2010; 21: 1335–1349.
Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 2009; 16: 398–410.
Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H . Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol 2008; 28: 3177–3189.
Vial C, Zuniga LM, Cabello-Verrugio C, Canon P, Fadic R, Brandan E . Skeletal muscle cells express the profibrotic cytokine connective tissue growth factor (CTGF/CCN2), which induces their dedifferentiation. J Cell Physiol 2008; 215: 410–421.
Watt KI, Judson R, Medlow P, Reid K, Kurth TB, Burniston JG et al. Yap is a novel regulator of C2C12 myogenesis. Biochem Biophys Res Commun 2010; 393: 619–624.
Chen HH, Maeda T, Mullett SJ, Stewart AF . Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis 2004; 39: 273–279.
Delacroix L, Moutier E, Altobelli G, Legras S, Poch O, Choukrallah MA et al. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol 2010; 30: 231–244.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515.
Anders S, Huber W . Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106.
Acknowledgements
We thank K Hnia and J Laporte for the gift of antibodies and advice. This work was supported by grants from the CNRS, the INSERM, the Université de Strasbourg, the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, the INCA and the AFM. ID is an ‘équipe labellisée’ of the Ligue Nationale contre le Cancer. AB was supported by a followship from the AFM. The mRNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE27845 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27845). The ChIP-chip data are are accessible through GEO Series accession number GSE29208.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by B Dynlacht
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Benhaddou, A., Keime, C., Ye, T. et al. Transcription factor TEAD4 regulates expression of Myogenin and the unfolded protein response genes during C2C12 cell differentiation. Cell Death Differ 19, 220–231 (2012). https://doi.org/10.1038/cdd.2011.87
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2011.87
Keywords
This article is cited by
-
Tec1, a member of the TEA transcription factors family, is involved in virulence and basidiocarp development in Ustilago maydis
International Microbiology (2022)
-
Activation of YAP regulates muscle fiber size in a PKC-dependent mechanism during chick in vitro myogenesis
Journal of Muscle Research and Cell Motility (2022)
-
Polymorphisms and genetic effects of PRLR, MOGAT1, MINPP1 and CHUK genes on milk fatty acid traits in Chinese Holstein
BMC Genetics (2019)
-
Cell-type dependent enhancer binding of the EWS/ATF1 fusion gene in clear cell sarcomas
Nature Communications (2019)
-
Identification of target genes in cardiomyopathy with fibrosis and cardiac remodeling
Journal of Biomedical Science (2018)