Abstract
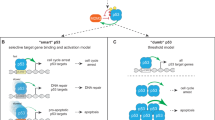
The p53 tumor suppressor responds to certain cellular stresses by inducing transcriptional programs that can lead to growth arrest or apoptosis. However, the molecular mechanisms responsible for choosing between these two cell fates are not well understood. Previous studies have suggested that p53 selectively activates proarrest target genes, due to the higher affinity of p53 for their promoters compared with proapoptotic genes. Here we show using microarray and chromatin immunoprecipitation that p53 binds to and transcriptionally activates both its proarrest and proapoptotic target genes proportionally to induced p53 expression levels. Further, we provide evidence that to trigger apoptosis, cells must overcome an apoptotic threshold, whose height is determined by expression levels of p53 and its targets, the duration of their expression and the cellular context. We demonstrate in multiple cells lines that below this threshold, expression levels of p53 and its targets were sufficient to induce arrest but not apoptosis. Above this threshold, p53 and its targets triggered extensive apoptosis. Moreover, lowering this threshold with inhibitors of antiapoptotic Bcl-2 family proteins sensitized cells to p53-induced apoptosis. These findings argue that agents that lower the apoptotic threshold should increase the efficacy of p53-mediated cancer therapy.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
Abbreviations
- ChIP:
-
chromatin immunoprecipitation
- dox:
-
doxycycline
- HMEC:
-
human mammary epithelial cells
- γH2AX:
-
phosphorylated form of H2AX
- PI:
-
propidium iodide
References
Vousden KH, Prives C . Blinded by the light: the growing complexity of p53. Cell 2009; 137: 413–431.
Horn HF, Vousden KH . Coping with stress: multiple ways to activate p53. Oncogene 2007; 26: 1306–1316.
Riley T, Sontag E, Chen P, Levine A . Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402–412.
Toledo F, Wahl GM . Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006; 6: 909–923.
Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP . Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 2009; 9: 862–873.
Cheok CF, Verma CS, Baselga J, Lane DP . Translating p53 into the clinic. Nat Rev Clin Oncol 2011; 8: 25–37.
Millar AW, Lynch KP . Rethinking clinical trials for cytostatic drugs. Nat Rev Cancer 2003; 3: 540–545.
Jackson JG, Post SM, Lozano G . Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. J Pathol 2010; 223: 127–137.
Murray-Zmijewski F, Slee EA, Lu X . A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol 2008; 9: 702–712.
Oren M . Decision making by p53: life, death and cancer. Cell Death Differ 2003; 10: 431–442.
Chen X, Ko LJ, Jayaraman L, Prives C . p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev 1996; 10: 2438–2451.
Macip S, Igarashi M, Berggren P, Yu J, Lee SW, Aaronson SA . Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol Cell Biol 2003; 23: 8576–8585.
Vousden KH, Lu X . Live or let die: the cell’s response to p53. Nat Rev Cancer 2002; 2: 594–604.
Kaeser MD, Iggo RD . Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci USA 2002; 99: 95–100.
Szak ST, Mays D, Pietenpol JA . Kinetics of p53 binding to promoter sites in vivo. Mol Cell Biol 2001; 21: 3375–3386.
Jackson JG, Pereira-Smith OM . P53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res 2006; 66: 8356–8360.
Kho PS, Wang Z, Zhuang L, Li Y, Chew JL, Ng HH et al. P53-regulated transcriptional program associated with genotoxic stress-induced apoptosis. J Biol Chem 2004; 279: 21183–21192.
Hernández-Vargas H, Ballestar E, Carmona-Saez P, von Kobbe C, Bañón-Rodríguez I et al. Transcriptional profiling of MCF7 breast cancer cells in response to 5-fluorouracil: relationship with cell cycle changes and apoptosis, and identification of novel targets of p53. Int J Cancer 2006; 119: 1164–1175.
Robinson M, Jiang P, Cui J, Li J, Wang Y, Swaroop M et al. Global genechip profiling to identify genes responsive to p53-induced growth arrest and apoptosis in human lung carcinoma cells. Cancer Biol Ther 2003; 2: 406–415.
Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev 2000; 14: 981–993.
Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene 2003; 22: 3645–3654.
Xia M, Knezevic D, Vassilev LT . P21 does not protect cancer cells from apoptosis induced by nongenotoxic p53 activation. Oncogene 2011; 30: 346–355.
Stampfer MR, Bartley JC . Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc Natl Acad Sci USA 1985; 82: 2394–2398.
Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA 2006; 103: 1888–1893.
Minemoto Y, Gannon J, Masutani M, Nakagama H, Sasagawa T, Inoue M et al. Characterization of Adriamycin-Induced G2 Arrest and Its Abrogation by Caffeine in FL-Amnion Cells with or without p53. Experimental Cell Research 2001; 262: 37–48.
Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI . Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer cell 2006; 10: 501–514.
Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S . [gamma]H2AX and cancer. Nat Rev Cancer 2008; 8: 957–967.
Kamada S, Kikkawa U, Tsujimoto Y, Hunter T . Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s). J Biol Chem 2005; 280: 857–860.
Mercer WE, Shields MT, Lin D, Appella E, Ullrich SJ . Growth suppression induced by wild-type p53 protein is accompanied by selective down-regulation of proliferating-cell nuclear antigen expression 1991 Proc Natl Acad Sci 88: 1958–1962.
Wu K, Jiang S-W, Couch FJ . P53 mediates repression of the BRCA2 promoter and down-regulation of BRCA2 mRNA and protein levels in response to DNA damage. J Biol Chem 2003; 278: 15652–15660.
Lazaro-Trueba I, Arias C, Silva A . Double bolt regulation of Rad51 by p53: a role for transcriptional repression. Cell Cycle 2006; 5: 1062–1065.
Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M . A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet 2002; 30: 315–320.
Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK . Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 2009; 459: 428–432.
Lowe SW, Cepero E, Evan G . Intrinsic tumour suppression. Nature 2004; 432: 307–315.
Hemann MT, Lowe SW . The p53–Bcl-2 connection. Cell Death Differ 2006; 13: 1256–1259.
Grinkevich VV, Nikulenkov F, Shi Y, Enge M, Bao W, Maljukova A et al. Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell 2009; 15: 441–453.
Pujals A, Renouf B, Robert A, Chelouah S, Hollville E, Wiels J . Treatment with a BH3 mimetic overcomes the resistance of latency III EBV (+) cells to p53-mediated apoptosis. Cell Death Dis 2011; 2: e184.
Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 2008; 68: 3421–3428.
Wang G, Nikolovska-Coleska Z, Yang CY, Wang R, Tang G, Guo J et al. Structure-based design of potent small-molecule inhibitors of anti-apoptotic Bcl-2 proteins. J Med Chem 2006; 49: 6139–6142.
Spencer Sabrina L, Sorger Peter K . Measuring and modeling apoptosis in single cells. Cell 2011; 144: 926–939.
Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H et al. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J Cell Biol 2004; 166: 839–851.
Polager S, Ginsberg D . P53 and E2f: partners in life and death. Nat Rev Cancer 2009; 9: 738–748.
Vousden KH, Lane DP . P53 in health and disease. Nat Rev Mol Cell Biol 2007; 8: 275–283.
Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer cell 2008; 14: 447–457.
Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA . Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol 2007; 9: 493–505.
Sewing A, Wiseman B, Lloyd AC, Land H . High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol 1997; 17: 5588–5597.
Sewing A, Wiseman B, Lloyd AC, Land H . High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol 1997; 17: 5588–5597.
Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H et al. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acutemyelogenous leukemia. Blood 2010; 116: 71–80.
Akiri G, Cherian MM, Vijayakumar S, Liu G, Bafico A, Aaronson SA . Wnt pathway aberrations including autocrine Wnt activation occur at high frequency in human non-small-cell lung carcinoma. Oncogene 2009; 28: 2163–2172.
Acknowledgements
We are grateful to Dr. Bo Zhao for his help with cloning procedures. Confocal laser scanning microscopy was performed at the Mount Sinai School of Medicine Microscopy Shared Resources Facility, supported with funding from NIH-NCI shared resources (Grant no. 5R24CA095823-04), National Science Foundation Major Research Instrumentation (Grant no. DBI-9724504) and NIH shared instrumentation (Grant no. 1 S10RR0 9145-01). We are also grateful to Dr. Huei-Chi Wen for his help with the confocal imaging analysis. We thank Luis Carvajal and Dr. Pierre-Jacques Hamard from Dr. James J Manfredi’s lab for sharing their ChIP assay expertise and providing us with the ChIP MDM2, p21 and PUMA primer sequences, Dr. Greg Khitrov in the Departmental Core Life Sciences Technology Laboratory of the Mount Sinai School of Medicine for performing the hybridization and processing of our Gene 1.1 ST Arrays, and Dr. Sam W Lee for helpful discussion. This study was supported by grants from the Breast Cancer Research Foundation, the Empire State Stem Cell Fund through New York State Department of Health Contract no. C024313 (SAA) and by P01 CA080058 from the National Cancer Institute (SAA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Empire State Stem Cell Board, the New York State Department of Health, the State of New York, the National Cancer Institute or the National Institutes of Health. Supplementary Information is available at Cell Death and Differentiation’s website.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Oren
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Kracikova, M., Akiri, G., George, A. et al. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ 20, 576–588 (2013). https://doi.org/10.1038/cdd.2012.155
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2012.155
Keywords
This article is cited by
-
Determinants of p53 DNA binding, gene regulation, and cell fate decisions
Cell Death & Differentiation (2024)
-
Emerging role and therapeutic implications of p53 in intervertebral disc degeneration
Cell Death Discovery (2023)
-
Formal verification confirms the role of p53 protein in cell fate decision mechanism
Theory in Biosciences (2023)
-
Partial p53 reactivation is sufficient to induce cancer regression
Journal of Experimental & Clinical Cancer Research (2022)
-
p53 partial loss-of-function mutations sensitize to chemotherapy
Oncogene (2022)