Abstract
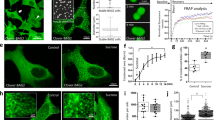
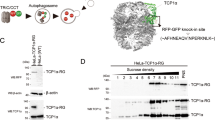
Eukaryotic cells are equipped with an efficient quality control system to selectively eliminate misfolded and damaged proteins, and organelles. Abnormal polypeptides that escape from proteasome-dependent degradation and aggregate in the cytosol can be transported via microtubules to inclusion bodies called ‘aggresomes’, where misfolded proteins are confined and degraded by autophagy. Here, we show that Type 2 transglutaminase (TG2) knockout mice display impaired autophagy and accumulate ubiquitinated protein aggregates upon starvation. Furthermore, p62-dependent peroxisome degradation is also impaired in the absence of TG2. We also demonstrate that, under cellular stressful conditions, TG2 physically interacts with p62 and they are localized in cytosolic protein aggregates, which are then recruited into autophagosomes, where TG2 is degraded. Interestingly, the enzyme's crosslinking activity is activated during autophagy and its inhibition leads to the accumulation of ubiquitinated proteins. Taken together, these data indicate that the TG2 transamidating activity has an important role in the assembly of protein aggregates, as well as in the clearance of damaged organelles by macroautophagy.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- TG2:
-
Type 2 transglutaminase
- Starv:
-
starvation
- CQ:
-
chloroquine
- 2- DG:
-
2-deoxy-D-glucose
- Rap:
-
rapamycin
- MG132:
-
Z-Leu-Leu-Leu-al
- VCP:
-
valosin-containing protein
- PMP70:
-
peroxisomal membrane protein
- HD:
-
Huntington's disease
- MEFs:
-
mouse embryonic fibroblast
References
Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR . Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000; 404: 770–774.
Yao TP . The role of ubiquitin in autophagy-dependent protein aggregate processing. Genes Cancer 2010; 1: 779–786.
Cyr DM, Hohfeld J, Patterson C . Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci 2002; 27: 368–375.
Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C . Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res 2010; 106: 463–478.
Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet 2003; 12: 749–757.
Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B . Aggregated and monomeric alpha-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J Biol Chem 2003; 278: 11753–11759.
Klionsky DJ, Emr SD . Autophagy as a regulated pathway of cellular degradation. Science 2000; 290: 1717–1721.
Klionsky DJ . Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8: 931–937.
Lamark T, Kirkin V, Dikic I, Johansen T . NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009; 8: 1986–1990.
Fesus L, Piacentini M . Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem Sci 2002; 27: 534–539.
Lai TS, Bielawska A, Peoples KA, Hannun YA, Greenberg CS . Sphingosylphosphocholine reduces the calcium ion requirement for activating tissue transglutaminase. J Biol Chem 1997; 272: 16295–16300.
Fesus L, Thomazy V, Autuori F, Ceru MP, Tarcsa E, Piacentini M . Apoptotic hepatocytes become insoluble in detergents and chaotropic agents as a result of transglutaminase action. FEBS Lett 1989; 245: 150–154.
Malorni W, Farrace MG, Rodolfo C, Piacentini M . Type 2 transglutaminase in neurodegenerative diseases: the mitochondrial connection. Curr Pharm Des 2008; 14: 278–288.
Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H . Mallory-Denk-bodies: lessons from keratincontaining hepatic inclusion bodies. Biochim Biophys Acta 2008; 1782: 764–774.
D’Eletto M, Farrace MG, Falasca L, Reali V, Oliverio S, Melino G et al. Transglutaminase 2 is involved in autophagosome maturation. Autophagy 2009; 5: 1145–1154.
Rossin F, D’Eletto M, Macdonald D, Farrace MG, Piacentini M . TG2 transamidating activity acts as a reostat controlling the interplay between apoptosis and autophagy. Amino Acids 2011. doi:10.1007/s00726-011-0899-x (in press).
Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y . In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15: 1101–1111.
Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina DL, Settembre C et al. Cystic fibrosis: a disorder with defective autophagy. Autophagy 2011; 7: 104–106.
Weidberg H, Shvets E, Elazar Z . Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80: 125–156.
Guthrie CR, Kraemer BC . Proteasome inhibition drives HDAC6-dependent recruitment of tau to aggresomes. J Mol Neurosci 2011; 45: 32–41.
Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J . Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA 2008; 105: 20567–20574.
Fahimi HD, Reinicke A, Sujatta M, Yokota S, Ozel M, Hartig F et al. The short- and long-term effects of bezafibrate in the rat. Ann NY Acad Sci 1982; 386: 111–135.
Islinger M, Luers GH, Li KW, Loos M, Volkl A . Rat liver peroxisomes after fibrate treatment: a survey using quantitative mass spectrometry. J Biol Chem 2007; 28: 23055–23069.
Dzhekova-Stojkova S, Bogdanska J, Stojkova Z . Peroxisome proliferators: their biological and toxicological effects. Clin Chem Lab Med 2001; 39: 468–474.
Ezaki J, Komatsu M, Yokota S, Ueno T, Kominami E . Method for monitoring pexophagy in mammalian cells. Methods Enzymol 2009; 452: 215–226.
Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M et al. Autophagy regulates lipid metabolism. Nature 2009; 458: 1131–1135.
Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell 2007; 25: 193–205.
Mastroberardino PG, Iannicola C, Nardacci R, Bernassola F, De Laurenzi V, Melino G et al. ‘Tissue’ transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington’s disease. Cell Death Differ 2002; 9: 873–880.
Biederbick A, Kern HF, Elsässer HP . Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol 1995; 66: 3–14.
Behrends C, Sowa ME, Gygi SP, Harper JW . Network organization of the human autophagy system. Nature 2010; 466: 68–76.
Hitomi K, Kitamura M, Sugimura Y . Preferred substrate sequences for transglutaminase 2: screening using a phage-displayed peptide library. Amino Acids 2009; 36: 619–624.
De Laurenzi V, Melino G . Gene disruption of tissue transglutaminase. Mol Cell Biol 2001; 21: 148–155.
Lewis MD, Pfeil J, Mueller AK . Continuous oral chloroquine as a novel route for Plasmodium prophylaxis and cure in experimental murine models. BMC Res Notes 2011; 4: 262–266.
Acknowledgements
The authors would like to thank Dr. P Mattioli from the Centre of Avanced Microscopy (CAM), Department of Biology, University of Rome Tor Vergata, for her skillful assistance in the use of the facility. This work was supported by grants from CHDI Foundation Inc. (USA), Compagnia di San Paolo, the Ministry of Health of Italy ‘Ricerca Corrente’ and ‘Ricerca Finalizzata’, and AIRC. The support of the EU grant ‘Apo-Sys’ and ‘Transpath’ Marie Curie project to MP is also acknowledged. FC is supported by grants from AIRC, Telethon and FISM.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by RA Knight
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
D'Eletto, M., Farrace, M., Rossin, F. et al. Type 2 transglutaminase is involved in the autophagy-dependent clearance of ubiquitinated proteins. Cell Death Differ 19, 1228–1238 (2012). https://doi.org/10.1038/cdd.2012.2
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2012.2
Keywords
This article is cited by
-
Type 2 transglutaminase in the nucleus: the new epigenetic face of a cytoplasmic enzyme
Cellular and Molecular Life Sciences (2023)
-
Non-alcoholic fatty liver disease severity is modulated by transglutaminase type 2
Cell Death & Disease (2018)
-
The roles of PKCs in regulating autophagy
Journal of Cancer Research and Clinical Oncology (2018)
-
Polyamines: therapeutic perspectives in oxidative stress and inflammatory diseases
Amino Acids (2017)
-
Transglutaminase 2, a double face enzyme
Amino Acids (2017)