Abstract
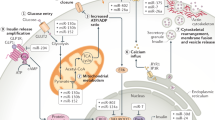
Our previous studies have demonstrated that stable microRNAs (miRNAs) in mammalian serum and plasma are actively secreted from tissues and cells and can serve as a novel class of biomarkers for diseases, and act as signaling molecules in intercellular communication. Here, we report the surprising finding that exogenous plant miRNAs are present in the sera and tissues of various animals and that these exogenous plant miRNAs are primarily acquired orally, through food intake. MIR168a is abundant in rice and is one of the most highly enriched exogenous plant miRNAs in the sera of Chinese subjects. Functional studies in vitro and in vivo demonstrated that MIR168a could bind to the human/mouse low-density lipoprotein receptor adapter protein 1 (LDLRAP1) mRNA, inhibit LDLRAP1 expression in liver, and consequently decrease LDL removal from mouse plasma. These findings demonstrate that exogenous plant miRNAs in food can regulate the expression of target genes in mammals.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297.
He L, Hannon GJ . MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522–531.
Calin GA, Croce CM . MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857–866.
Esquela-Kerscher A, Slack FJ . Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259–269.
Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18:997–1006.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008; 105:10513–10518.
Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One 2008; 3:e3148.
Li LM, Hu ZB, Zhou ZX, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res 2010; 70:9798–9807.
Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009; 58:1375–1381.
Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE . The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol 2009; 112:55–59.
Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA 2009; 106:4402–4407.
Zhang C, Wang C, Chen X, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem 2010; 56:1871–1879.
Cocucci E, Racchetti G, Meldolesi J . Shedding microvesicles: artefacts no more. Trends Cell Biol 2009; 19:43–51.
Thery C, Zitvogel L, Amigorena S . Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002; 2:569–579.
Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010; 39:133–144.
Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10:1470–1476.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO . Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654–659.
Collino F, Deregibus MC, Bruno S, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One 2010; 5:e 11803.
Meckes DG Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N . Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA 2010; 107:20370–20375.
Ogawa R, Tanaka C, Sato M, et al. Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem Biophys Res Commun 2010; 398:723–729.
Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 2010; 107:6328–6333.
Yu B, Yang Z, Li J, et al. Methylation as a crucial step in plant microRNA biogenesis. Science 2005; 307:932–935.
Grimson A, Srivastava M, Fahey B, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 2008; 455:1193–1197.
Hartig JV, Forstemann K . Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res 2011; 39:3836–3851.
Gazzani S, Li M, Maistri S, et al. Evolution of MIR168 paralogs in Brassicaceae. BMC Evol Biol 2009; 9:62.
Jia X, Mendu V, Tang G . An array platform for identification of stress-responsive microRNAs in plants. Methods Mol Biol 2010; 639:253–269.
Chen X . MicroRNA biogenesis and function in plants. FEBS Lett 2005; 579:5923–5931.
Du T, Zamore PD . microPrimer: the biogenesis and function of microRNA. Development 2005; 132:4645–4652.
Garcia CK, Wilund K, Arca M, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science 2001; 292:1394–1398.
Huang S, Wu S, Ding J, et al. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res 2010; 38:7211–7218.
Qin W, Shi Y, Zhao B, et al. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One 2010; 5:e9429.
Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I . MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008; 455:1124–1128.
Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O . Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 2009; 11:1143–1149.
Goldstein JL, Brown MS . The low-density lipoprotein pathway and its relation to atherosclerosis. Annu Rev Biochem 1977; 46:897–930.
Wilund KR, Yi M, Campagna F, et al. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum Mol Genet 2002; 11:3019–3030.
Zuliani G, Arca M, Signore A, et al. Characterization of a new form of inherited hypercholesterolemia: familial recessive hypercholesterolemia. Arterioscler Thromb Vasc Biol 1999; 19:802–809.
Mallory AC, Vaucheret H . ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep 2009; 10:521–526.
Vaucheret H, Mallory AC, Bartel DP . AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 2006; 22:129–136.
Vaucheret H, Vazquez F, Crete P, Bartel DP . The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 2004; 18:1187–1197.
Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST . Binding and cleavage specificities of human Argonaute2. J Biol Chem 2009; 284:26017–26028.
Brown BD, Naldini L . Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet 2009; 10:578–585.
Brown BD, Gentner B, Cantore A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 2007; 25:1457–1467.
Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005; 33:e179.
Neilson JR, Zheng GX, Burge CB, Sharp PA . Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev 2007; 21:578–589.
Bissels U, Wild S, Tomiuk S, et al. Absolute quantification of microRNAs by using a universal reference. RNA 2009; 15:2375–2384.
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T . MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007; 133:647–658.
Shimizu S, Takehara T, Hikita H, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol 2010; 52:698–704.
Tomimaru Y, Eguchi H, Nagano H, et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer 2010; 103:1617–1626.
Zhang J, Yang Y, Yang T, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer 2010; 103:1215–1220.
Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P . Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005; 309:1577–1581.
Roney JK, Khatibi PA, Westwood JH . Cross-species translocation of mRNA from host plants into the parasitic plant dodder. Plant Physiol 2007; 143:1037–1043.
Chen X, Li Q, Wang J, et al. Identification and characterization of novel amphioxus microRNAs by Solexa sequencing. Genome Biol 2009; 10:R78.
Jiang J, Lee EJ, Gusev Y, Schmittgen TD . Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 2005; 33:5394–5403.
Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K . The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 2007; 14:349–350.
Smalheiser NR, Torvik VI . Complications in mammalian microRNA target prediction. Methods Mol Biol 2006; 342:115–127.
Acknowledgements
We thank Dr Daniel Wu (Yale University, USA) and Dr Zheng-Gang Liu (Cancer Research Institute, NIH, USA) for critical reading of the manuscript and discussion of data. This work was supported by grants from the National Natural Science Foundation of China (90813035, 30890044, 30771036, 30772484, 30725008, 30890032, 31071232, 31000323, and 90608010), the National Basic Research Program of China (973 Program 2006CB503909, 2007CB815701, 2007CB815703, 2007CB815705, and 2007CB815804; 863 Program 2006AA02Z177 and 2006AA10A121), and the Natural Science Foundation of Jiangsu Province (BK2008021).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
The level and sequence of plant miRNAs in human serum. (PDF 40 kb)
Supplementary information, Table S2
The level and sequence of plant miRNAs in calf serum. (PDF 37 kb)
Supplementary information, Table S3
The level of plant miRNAs in chow diet, rice, Chinese cabbage, wheat, and potato. (PDF 37 kb)
Supplementary information, Table S4
Potential mammalian genes identified as MIR168a targets. (PDF 42 kb)
Supplementary information, Table S5
The fundamental ingredients of the chow diet and fresh rice. (PDF 9 kb)
Supplementary information, Figure S1
The detection and analysis of plant miRNAs in serum samples of mammals. (PDF 116 kb)
Supplementary information, Figure S2
A quantitative analysis of plant miRNAs in mouse serum and various organs as well as the uptake of various forms of MIR168a by C57BL/6J mice. (PDF 164 kb)
Supplementary information, Figure S3
Targeting of LDLRAP1 by MIR168a. (PDF 240 kb)
Supplementary information, Figure S4
An analysis of the MIR168a levels in MVs from 293T or Caco-2 cells transfected with 20 pmol/105 MIR168a and in HepG2 cells after treatment with MVs from 293T or Caco-2 cells. (PDF 108 kb)
Supplementary information, Figure S5
The levels of MIR168a in various mouse organs after chow diet or fresh rice feeding for sustained periods (n = 8). (PDF 164 kb)
Supplementary information, Figure S6
The effects of LDLRAP1 siRNA on the levels of mouse liver LDLRAP1 and mouse plasma LDL-cholesterol, cholesterol, and triglycerides. (PDF 132 kb)
Supplementary information, Figure S7
The effects of the rice-derived MIR168a and supplemental mature miR-150 in chow diet on mouse plasma cholesterol, lipoprotein, and triglycerides and liver c-Myb protein levels. (PDF 176 kb)
Rights and permissions
About this article
Cite this article
Zhang, L., Hou, D., Chen, X. et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 22, 107–126 (2012). https://doi.org/10.1038/cr.2011.158
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2011.158
Keywords
This article is cited by
-
Alfalfa xeno-miR159a regulates bovine mammary epithelial cell proliferation and milk protein synthesis by targeting PTPRF
Scientific Reports (2024)
-
Novel plant-derived exosome-like nanovesicles from Catharanthus roseus: preparation, characterization, and immunostimulatory effect via TNF-α/NF-κB/PU.1 axis
Journal of Nanobiotechnology (2023)
-
Employing in silico investigations to determine the cross-kingdom approach for Curcuma longa miRNAs and their human targets
Beni-Suef University Journal of Basic and Applied Sciences (2023)
-
Plant miRNA osa-miR172d-5p suppressed lung fibrosis by targeting Tab1
Scientific Reports (2023)
-
Are herbal sRNAs really novel precision medicines?
Science China Life Sciences (2023)