Abstract
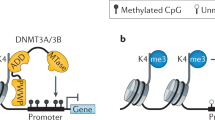
Cytosine methylation of genomic DNA controls gene expression and maintains genome stability. How a specific DNA sequence is targeted for methylation by a methyltransferase is largely unknown. Here, we show that histone H3 tails lacking lysine 4 (K4) methylation function as an allosteric activator for methyltransferase Dnmt3a by binding to its plant homeodomain (PHD). In vitro, histone H3 peptides stimulated the methylation activity of Dnmt3a up to 8-fold, in a manner reversely correlated with the level of K4 methylation. The biological significance of allosteric regulation was manifested by molecular modeling and identification of key residues in both the PHD and the catalytic domain of Dnmt3a whose mutations impaired the stimulation of methylation activity by H3 peptides but not the binding of H3 peptides. Significantly, these mutant Dnmt3a proteins were almost inactive in DNA methylation when expressed in mouse embryonic stem cells while their recruitment to genomic targets was unaltered. We therefore propose a two-step mechanism for de novo DNA methylation – first recruitment of the methyltransferase probably assisted by a chromatin- or DNA-binding factor, and then allosteric activation depending on the interaction between Dnmt3a and the histone tails – the latter might serve as a checkpoint for the methylation activity.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Jaenisch R, Bird A . Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33 Suppl:245–254.
Bestor TH . The DNA methyltransferases of mammals. Hum Mol Genet 2000; 9:2395–2402.
Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010; 363:2424–2433.
Ciccone DN, Su H, Hevi S, et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009; 461:415–418.
Dennis K, Fan T, Geiman T, Yan Q, Muegge K . Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev 2001; 15:2940–2944.
Fan Y, Nikitina T, Zhao J, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 2005; 123:1199–1212.
Cheng X, Blumenthal RM . Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry 2010; 49:2999–3008.
Li E . Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 2002; 3:662–673.
Tamaru H, Selker EU . A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 2001; 414:277–283.
Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 2003; 13:1192–1200.
Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 2008; 30:755–766.
Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454:766–770.
Ooi SK, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007; 448:714–717.
Otani J, Nankumo T, Arita K, et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep 2009; 10:1235–1241.
Zhang Y, Jurkowska R, Soeroes S, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 2010; 38:4246–4253.
Hu JL, Zhou BO, Zhang RR, et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci USA 2009; 106:22187–22192.
Cedar H, Bergman Y . Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009; 10:295–304.
Dhayalan A, Rajavelu A, Rathert P, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 2010; 285:26114–26120.
Ge YZ, Pu MT, Gowher H, et al. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J Biol Chem 2004; 279:25447–25454.
Chen T, Tsujimoto N, Li E . The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol 2004; 24:9048–9058.
Hsieh CL . In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol Cell Biol 1999; 19:8211–8218.
Chen T, Ueda Y, Dodge JE, Wang Z, Li E . Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 2003; 23:5594–5605.
Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X . Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 2007; 449:248–251.
Tsumura A, Hayakawa T, Kumaki Y, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 2006; 11:805–814.
Jones PA, Liang G . Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 2009; 10:805–811.
Dong KB, Maksakova IA, Mohn F, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 2008; 27:2691–2701.
Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y . G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 2008; 27:2681–2690.
Epsztejn-Litman S, Feldman N, Abu-Remaileh M, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 2008; 15:1176–1183.
Meilinger D, Fellinger K, Bultmann S, et al. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009; 10:1259–1264.
Di Croce L, Raker VA, Corsaro M et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 2002; 295:1079–1082.
Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39:457–466.
Chodavarapu RK, Feng S, Bernatavichute YV, et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010; 466:388–392.
Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 2006; 8:188–194.
Fuhrmann G, Chung AC, Jackson KJ, et al. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev Cell 2001; 1:377–387.
Xie ZH, Huang YN, Chen ZX, et al. Mutations in DNA methyltransferase DNMT3B in ICF syndrome affect its regulation by DNMT3L. Hum Mol Genet 2006; 15:1375–1385.
Roth M, Jeltsch A . Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol Chem 2000; 381:269–272.
Ramsahoye BH . Measurement of genome wide DNA methylation by reversed-phase high-performance liquid chromatography. Methods 2002; 27:156–161.
Li JY, Pu MT, Hirasawa R, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol 2007; 27:8748–8759.
Okano M, Bell DW, Haber DA, Li E . DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99:247–257.
Shi X, Hong T, Walter KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006; 442:96–99.
Li H, Ilin S, Wang W, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006; 442:91–95.
Hu YG, Hirasawa R, Hu JL, et al. Regulation of DNA methylation activity through Dnmt3L promoter methylation by Dnmt3 enzymes in embryonic development. Hum Mol Genet 2008; 17:2654–2664.
Acknowledgements
We thank Gerd Pfeifer (Beckman Research Institute of City of Hope) and Taiping Chen (Novartis Institutes for Biomedical Research) for helpful discussions and Jiemin Wong (East China Normal University) for critical reading of the manuscript. We are grateful to Masaki Okano (RIKEN Center for Developmental Biology) for providing Dnmt triple knockout ES cells. This work was supported by grants from the Ministry of Science and Technology of China (2005CB522400 and 2006CB943900), the National Natural Science Foundation of China (30730059) and the Shanghai Municipal Government (08dj1400501, 0852S11223) to GLX.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
The Dnmt3a PHD domain interacts with the catalytic domain. (PDF 83 kb)
Supplementary information, Figure S2
Measurement of binding affinity of Flag-Dnmt3a toward DNA. (PDF 98 kb)
Supplementary information, Figure S3
Coomassie staining of Flag-tagged full-length Dnmt3a proteins purified from transiently transfected HEK293T cells and GST-fusion proteins of 3aPHD purified from E. coli BL21 (DE3). (PDF 169 kb)
Supplementary information, Figure S4
Functional analysis of Dnmt3a D308A and M325W mutants. (PDF 146 kb)
Supplementary information, Figure S5
Southern hybridization analysis of the methylation activity of transiently transfected Dnmt3a interface mutants. (PDF 121 kb)
Supplementary information, Figure S6
Expression and distribution of Dnmt3a Q304A and R580A mutants in mouse ES cells. (PDF 160 kb)
Supplementary information, Figure S7
Methylation analysis at the Oct4 and Sox30 promoters in ES cells ectopically expressing wild-type and mutant Dnmt3a. (PDF 146 kb)
Supplementary Table 1.
Mutational analysis of residues on the interfaces of the PHD and catalytic domains of Dnmt3a. (PDF 90 kb)
Supplementary information, Data S1
Materials and Methods (PDF 61 kb)
Rights and permissions
About this article
Cite this article
Li, BZ., Huang, Z., Cui, QY. et al. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res 21, 1172–1181 (2011). https://doi.org/10.1038/cr.2011.92
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2011.92
Keywords
This article is cited by
-
Regulation, functions and transmission of bivalent chromatin during mammalian development
Nature Reviews Molecular Cell Biology (2023)
-
Mechanisms of chromatin-based epigenetic inheritance
Science China Life Sciences (2022)
-
Enhancer-associated H3K4 methylation safeguards in vitro germline competence
Nature Communications (2021)
-
E2F6 initiates stable epigenetic silencing of germline genes during embryonic development
Nature Communications (2021)
-
DNMT3A reads and connects histone H3K36me2 to DNA methylation
Protein & Cell (2020)