Abstract
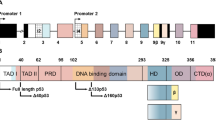
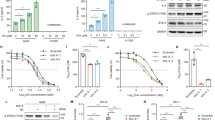
Alkylating agents induce genome-wide base damage, which is repaired mainly by N-methylpurine DNA glycosylase (MPG). An elevated expression of MPG in certain types of tumor cells confers higher sensitivity to alkylation agents because MPG-induced apurinic/apyrimidic (AP) sites trigger more strand breaks. However, the determinant of drug sensitivity or insensitivity still remains unclear. Here, we report that the p53 status coordinates with MPG to play a pivotal role in such process. MPG expression is positive in breast, lung and colon cancers (38.7%, 43.4% and 25.3%, respectively) but negative in all adjacent normal tissues. MPG directly binds to the tumor suppressor p53 and represses p53 activity in unstressed cells. The overexpression of MPG reduced, whereas depletion of MPG increased, the expression levels of pro-arrest gene downstream of p53 including p21, 14-3-3σ and Gadd45 but not proapoptotic ones. The N-terminal region of MPG was specifically required for the interaction with the DNA binding domain of p53. Upon DNA alkylation stress, in p53 wild-type tumor cells, p53 dissociated from MPG and induced cell growth arrest. Then, AP sites were repaired efficiently, which led to insensitivity to alkylating agents. By contrast, in p53-mutated cells, the AP sites were repaired with low efficacy. To our knowledge, this is the first direct evidence to show that a DNA repair enzyme functions as a selective regulator of p53, and these findings provide new insights into the functional linkage between MPG and p53 in cancer therapy.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Vousden KH, Lu X . Live or let die: the cell's response to p53. Nat Rev Cancer 2002; 2:594–604.
Kern SE, Kinzler KW, Bruskin A, et al. Identification of p53 as a sequence-specific DNA-binding protein. Science 1991; 252:1708–1711.
Harris SL, Levine AJ . The p53 pathway: positive and negative feedback loops. Oncogene 2005; 24:2899–2908.
Dumaz N, Meek DW . Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J 1999; 18:7002–7010.
Tian C, Xing G, Xie P, et al. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 2009; 11:580–591.
Sullivan A, Lu X . ASPP: a new family of oncogenes and tumour suppressor genes. Br J Cancer 2007; 96:196–200.
Tanaka T, Ohkubo S, Tatsuno I, Prives C . hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell 2007; 130:638–650.
Das S, Raj L, Zhao B, et al. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 2007; 130:624–637.
Schnack C, Hengerer B, Gillardon F . Identification of novel substrates for Cdk5 and new targets for Cdk5 inhibitors using high-density protein microarrays. Proteomics 2008; 8:1980–1986.
Samson L, Derfler B, Boosalis M, Call K . Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci USA 1991; 88:9127–9131.
Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW . The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res 2005; 65:6394–6400.
Fishel ML, He Y, Smith ML, Kelley MR . Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res 2007; 13:260–267.
Klapacz J, Lingaraju GM, Guo HH, et al. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Mol Cell 2010; 37:843–853.
Coquerelle T, Dosch J, Kaina B . Overexpression of N-methylpurine-DNA glycosylase in Chinese hamster ovary cells renders them more sensitive to the production of chromosomal aberrations by methylating agents--a case of imbalanced DNA repair. Mutat Res 1995; 336:9–17.
Posnick LM, Samson LD . Imbalanced base excision repair increases spontaneous mutation and alkylation sensitivity in Escherichia coli. J Bacteriol 1999; 181:6763–6771.
Gillen CD, Walmsley RS, Prior P, Andrews HA, Allan RN . Ulcerative colitis and Crohn's disease: a comparison of the colorectal cancer risk in extensive colitis. Gut 1994; 35:1590–1592.
Hofseth LJ, Saito S, Hussain SP, et al. Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proc Natl Acad Sci USA 2003; 100:143–148.
Paik J, Duncan T, Lindahl T, Sedgwick B . Sensitization of human carcinoma cells to alkylating agents by small interfering RNA suppression of 3-alkyladenine-DNA glycosylase. Cancer Res 2005; 65:10472–10477.
Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T . Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell 1998; 95:249–258.
Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T . Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA 2000; 97:13573–13578.
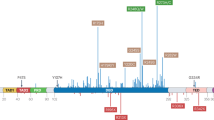
Berglind H, Pawitan Y, Kato S, Ishioka C, Soussi T . Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol Ther 2008; 7:699–708.
Hollstein M, Sidransky D, Vogelstein, B, Harris CC . p53 mutations in human cancers. Science 1991; 253:49–53.
Weisz L, Oren M, Rotter V . Transcription regulation by mutant p53. Oncogene 2007; 26:2202–2211.
Watanabe S, Ichimura T, Fujita N, et al. Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin. Proc Natl Acad Sci USA 2003; 100:12859–12864.
Lee CY, Delaney JC, Kartalou M, et al. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG). Biochemistry 2009; 48:1850–1861.
Adhikari S, Toretsky JA, Yuan L, Roy R . Magnesium, essential for base excision repair enzymes, inhibits substrate binding of N-methylpurine-DNA glycosylase. J Biol Chem 2006; 281:29525–29532.
Wood RD, Mitchell M, Sgouros J, Lindahl T . Human DNA repair genes. Science 2001; 291:1284–1289.
Tentori L, Graziani G . Pharmacological strategies to increase the antitumor activity of methylating agents. Curr Med Chem 2002; 9:1285–1301.
Rinne M, Caldwell D, Kelley MR . Transient adenoviral N-methylpurine DNA glycosylase overexpression imparts chemotherapeutic sensitivity to human breast cancer cells. Mol Cancer Ther 2004; 3:955–967.
Chou WC, Wang HC, Wong FH, et al. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J 2008; 27:3140–3150.
Nakamura J, Asakura S, Hester SD, de Murcia G, Caldecott KW, Swenberg JA . Quantitation of intracellular NAD(P)H can monitor an imbalance of DNA single strand break repair in base excision repair deficient cells in real time. Nucleic Acids Res 2003; 31:e104.
Swain U, Subba Rao K . Study of DNA damage via the comet assay and base excision repair activities in rat brain neurons and astrocytes during aging. Mech Ageing Dev 2011; 132:374–381.
Olive PL, Banath JP . The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006; 1:23–29.
Kow YW, Dare A . Detection of abasic sites and oxidative DNA base damage using an ELISA-like assay. Methods 2000; 22:164–169.
Bapat A, Glass LS, Luo M, et al. Novel small-molecule inhibitor of apurinic/apyrimidinic endonuclease 1 blocks proliferation and reduces viability of glioblastoma cells. J Pharmacol Exp Ther 2010; 334:988–998.
Drablos F, Feyzi E, Aas PA, et al. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004; 3:1389–1407.
Labahn J, Scharer OD, Long A, et al. Structural basis for the excision repair of alkylation-damaged DNA. Cell 1996; 86:321–329.
Dittmer D, Pati S, Zambetti G, et al. Gain of function mutations in p53. Nat Genet 1993; 4:42–46.
Hofseth LJ, Khan MA, Ambrose M, et al. The adaptive imbalance in base excision–repair enzymes generates microsatellite instability in chronic inflammation. J Clin Invest 2003; 112:1887–1894.
Longley DB, Harkin DP, Johnston PG . 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003; 3:330–338.
Miao F, Bouziane M, Dammann R, et al. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J Biol Chem 2000; 275:28433–28438.
Rui Y, Xu Z, Lin S, et al. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J 2004; 23:4583–4594.
Mi B, Wang X . Bai Y, et al. Beta-catenin expression is altered in dysplastic and nondysplastic aberrant crypt foci of human colon. Appl Immunohistochem Mol Morphol 2009; 17:294–300.
Abner CW, Lau AY, Ellenberger T . Bloom LB Base excision and DNA binding activities of human alkyladenine DNA glycosylase are sensitive to the base paired with a lesion. J Biol Chem 2001; 276:13379–13387.
Acknowledgements
We thank Drs Shengcai Lin (Xiamen University), Bert Vogelstein (Johns Hopkins Oncology Center) and Yue Xiong (University of North Carolina) for providing materials. The study was supported by the National Basic Research Programs (2011CB910802, 2012CB910702, 2010CB912202), and National Natural Science Foundation Projects (31071144, 31125010).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
MPG interacts with p53 and inhibits p53 activity in HEK293 cells. (PDF 19 kb)
Supplementary information, Figure S2
p53 has no significant influence on MPG-mediated excision of Hx. (PDF 17 kb)
Supplementary information, Figure S3
MPG selectively downregulates the expression of pro-arrest p53 target genes in HEK293 cells. (PDF 15 kb)
Supplementary information, Data S1
Experimental Protocols (PDF 22 kb)
Supplementary information, Table S1
Primer sequences used for qPCR assays. (PDF 24 kb)
Rights and permissions
About this article
Cite this article
Song, S., Xing, G., Yuan, L. et al. N-methylpurine DNA glycosylase inhibits p53-mediated cell cycle arrest and coordinates with p53 to determine sensitivity to alkylating agents. Cell Res 22, 1285–1303 (2012). https://doi.org/10.1038/cr.2012.107
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2012.107
Keywords
This article is cited by
-
Early onset senescence and cognitive impairment in a murine model of repeated mTBI
Acta Neuropathologica Communications (2021)
-
Molecular characterization of Plasmodium falciparum DNA-3-methyladenine glycosylase
Malaria Journal (2020)
-
Mapping genetic alterations causing chemoresistance in cancer: identifying the roads by tracking the drivers
Oncogene (2013)