Abstract
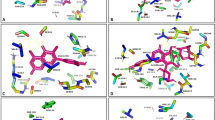
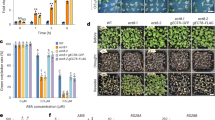
PYR1/PYL/RCAR family proteins (PYLs) are well-characterized abscisic acid (ABA) receptors. Among the 14 PYL members in Arabidopsis thaliana, PYL13 is ABA irresponsive and its function has remained elusive. Here, we show that PYL13 selectively inhibits the phosphatase activity of PP2CA independent of ABA. The crystal structure of PYL13-PP2CA complex, which was determined at 2.4 Å resolution, elucidates the molecular basis for the specific recognition between PP2CA and PYL13. In addition to the canonical interactions between PYLs and PP2Cs, an extra interface is identified involving an element in the vicinity of a previously uncharacterized CCCH zinc-finger (ZF) motif in PP2CA. Sequence blast identified another 56 ZF-containing PP2Cs, all of which are from plants. The structure also reveals the molecular determinants for the ABA irresponsiveness of PYL13. Finally, biochemical analysis suggests that PYL13 may hetero-oligomerize with PYL10. These two PYLs antagonize each other in their respective ABA-independent inhibitions of PP2Cs. The biochemical and structural studies provide important insights into the function of PYL13 in the stress response of plant and set up a foundation for future biotechnological applications of PYL13.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Fedoroff NV . Cross-talk in abscisic acid signaling. Sci STKE 2002; 2002:re10.
Finkelstein R, Reeves W, Ariizumi T, Steber C . Molecular aspects of seed dormancy. Annu Rev Plant Biol 2008; 59:387–415.
Schroeder JI, Nambara E . A quick release mechanism for abscisic acid. Cell 2006; 126:1023–1025.
Zhu JK . Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 2002; 53:247–273.
Ma Y, Szostkiewicz I, Korte A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009; 324:1064–1068.
Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009; 324:1068–1071.
Fujii H, Chinnusamy V, Rodrigues A, et al. In vitro reconstitution of an abscisic acid signaling pathway. Nature 2009; 462:660–664.
Santiago J, Rodrigues A, Saez A, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 2009; 60:575–588.
Xie T, Ren R, Zhang YY, et al. Molecular mechanism for inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by protein phosphatase ABI1. J Biol Chem 2012; 287:794–802.
Soon FF, Ng LM, Zhou XE, et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012; 335:85–88.
Yin P, Fan H, Hao Q, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 2009; 16:1230–1236.
Melcher K, Ng LM, Zhou XE, et al. A gate-latch-lock mechanism for hormone signaling by abscisic acid receptors. Nature 2009; 462:602–608.
Hao Q, Yin P, Li W, et al. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell 2011; 42:662–672.
Miyazono K, Miyakawa T, Sawano Y, et al. Structural basis of abscisic acid signaling. Nature 2009; 462:609–614.
Mosquna A, Peterson FC, Park SY, Lozano-Juste J, Volkman BF, Cutler SR . Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc Natl Acad Sci USA 2011; 108:20838–20843.
Heimovaara-Dijkstra S, Testerink C, Wang M . Mitogen-activated protein kinase and abscisic acid signal transduction. Results Probl Cell Differ 2000; 27:131–144.
Yuan XQ, Yin P, Hao Q, Yan CY, Wang JW, Yan NE . Single amino acid alteration between valine and isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2. J Biol Chem 2010; 285:28953–28958.
Melcher K, Xu Y, Ng LM, et al. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol 2010; 17:1102–1108.
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR . Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 2010; 61: 651–679.
Melcher K, Zhou XE, Xu HE . Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr Opin Struct Biol 2010; 20:722–729
Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M . Structure and function of abscisic acid receptors. Trends Plant Sci 2013; 18:259–266.
Gonzalez-Guzman M, Pizzio GA, Antoni R, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012; 24:2483–2496.
Sun D, Wang H, Wu M, Zang J, Wu F, Tian C . Crystal structures of the Arabidopsis thaliana abscisic acid receptor PYL10 and its complex with abscisic acid. Biochem Biophys Res Commun 2012; 418:122–127.
Shi Y . Serine/threonine phosphatases: mechanism through structure. Cell 2009; 139:468–484.
Hao Q, Yin P, Yan C, et al. Functional mechanism of the ABA agonist pyrabactin. J Biol Chem 2010; 285:28946–28952.
Yoshida T, Nishimura N, Kitahata N, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 2006; 140:115–126.
Lan WZ, Lee SC, Che YF, Jiang YQ, Luan S . Mechanistic analysis of AKT1 regulation by the CBL-IPK-P2CA interactions. Mol Plant 2011; 4:527–536.
Bhaskara GB, Nguyen TT, Verslues PE . Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 2012; 160:379–395.
Pomeranz M, Finer J, Jang JC . Putative molecular mechanisms underlying tandem CCCH zinc finger protein mediated plant growth, stress, and gene expression responses. Plant Signal Behav 2011; 6:647–651.
Otwinowski Z, Minor W . Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307–326.
Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr 1994; D50:760–763.
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ . Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674.
Emsley P, Cowtan K . Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132.
Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948–1954.
DeLano WL . The PyMOL Molecular Graphics System. Available at: http://www.pymolorg2002.
Acknowledgements
We thank J He and S Huang at Shanghai Synchrotron Radiation Facility (SSRF) for assistance. This work was supported by funds from the Ministry of Science and Technology (2011CB910501), the National Natural Science Foundation of China (31125009 and 91017011), and Tsinghua University. The research of Nieng Yan was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Sequence comparison of five Clade A PP2Cs from Arabidopsis thaliana. (PDF 581 kb)
Supplementary information, Figure S2
Sequence comparison of PP2CA with HAI1/2/3. (PDF 369 kb)
Supplementary information, Figure S3
Evolutionary relationships of ZF (zinc-finger motif)-containing PP2Cs. (PDF 384 kb)
Supplementary information, Figure S4
Structure-guided point mutations converted PYL13 into an ABA receptor. (PDF 92 kb)
Supplementary information, Figure S5
PYL13 and PYL10 form hetero-complex. (PDF 37 kb)
Supplementary information Table S1
Statistics of data collection and refinement (PDF 30 kb)
Supplementary information Table S2
ICP-AES (Inductively Coupled Plasma-Atomic Emission Spectrometry) analysis of the metal contents in PP2CA-PYL13 complex and PYL5 (PDF 34 kb)
Rights and permissions
About this article
Cite this article
Li, W., Wang, L., Sheng, X. et al. Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res 23, 1369–1379 (2013). https://doi.org/10.1038/cr.2013.143
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2013.143
Keywords
This article is cited by
-
Genome-wide identification of the PYL gene family of tea plants (Camellia sinensis) revealed its expression profiles under different stress and tissues
BMC Genomics (2023)
-
GhPYL9-5D and GhPYR1-3 A positively regulate Arabidopsis and cotton responses to ABA, drought, high salinity and osmotic stress
BMC Plant Biology (2023)
-
Hormonal and Transcriptomic Analysis Reveal Gibberellin-Induced Fruit Set in Jujube (Ziziphus jujuba Mill.)
Plant Molecular Biology Reporter (2022)
-
Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure
Nature Communications (2021)
-
Influence of virus–host interactions on plant response to abiotic stress
Plant Cell Reports (2021)