Abstract
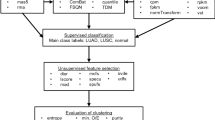
Lung squamous cell carcinoma (SCC) is one of the major subtypes of lung cancer. Our current knowledge of oncogenic drivers in this specific subtype of lung cancer is largely limited compared with lung adenocarcinoma (ADC). Through exon array analyses, molecular analyses and functional studies, we here identify the TRA2B-DNAH5 fusion as a novel oncogenic driver in lung SCC. We found that this gene fusion occurs exclusively in lung SCC (3.1%, 5/163), but not in lung ADC (0/119). Through mechanistic studies, we further revealed that this TRA2B-DNAH5 fusion promotes lung SCC malignant progression through regulating a SIRT6-ERK1/2-MMP1 signaling axis. We show that inhibition of ERK1/2 activation using selumetinib efficiently inhibits the growth of lung SCC with TRA2B-DNAH5 fusion expression. These findings improve our current knowledge of oncogenic drivers in lung SCC and provide a potential therapeutic strategy for lung SCC patients with TRA2B-DNAH5 fusion.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Siegel R, Ma J, Zou Z, Jemal A . Cancer statistics. CA Cancer J Clin 2014; 64:9–29.
Herbst RS, Lippman SM . Molecular signatures of lung cancer — toward personalized therapy. N Engl J Med 2007; 356:76–78.
Riely GJ, Marks J, Pao W . KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 2009; 6:201–205.
Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 2004; 101:13306–13311.
Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304:1497–1500.
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350:2129–2139.
Ramos AH, Dutt A, Mermel C, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther 2009; 8:2042–2050.
Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov 2011; 1:78–89.
Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448:561–566.
Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007; 131:1190–1203.
Li F, Feng Y, Fang R, et al. Identification of RET gene fusion by exon array analyses in “pan-negative” lung cancer from never smokers. Cell Res 2012; 22:928–931.
Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012; 18:382–384.
Ju YS, Lee WC, Shin JY, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res 2012; 22:436–445.
Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012; 18:375–377.
Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012; 18:378–381.
Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013; 19:1469–1472.
Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013; 3:636–647.
Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014; 4:415–422.
Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013; 368:2385–2394.
Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014; 371:2167–2177.
Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014; 371:1963–1971.
Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013; 3:630–635.
Li F, Feng Y, Fang R, et al. Identification of RET gene fusion by exon array analyses in “pan-negative” lung cancer from never smokers. Cell Res 2012; 22:928–931.
Pao W, Girard N . New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011; 12:175–180.
Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010; 28:4616–4620.
Li C, Fang R, Sun Y, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One 2011; 6:e28204.
Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res 2009; 7:1466–1476.
Seegar TC, Eller B, Tzvetkova-Robev D, et al. Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol Cell 2010; 37:643–655.
Foley CJ, Kuliopulos A . Mouse matrix metalloprotease-1a (Mmp1a) gives new insight into MMP function. J Cell Physiol 2014; 229:1875–1880.
Kim CH, Park YG, Noh SH, Kim YK . PGE2 induces the gene expression of bone matrix metalloproteinase-1 in mouse osteoblasts by cAMP-PKA signaling pathway. Int J Biochem Cell Biol 2005; 37:375–385.
Sokolova O, Vieth M, Naumann M . Protein kinase C isozymes regulate matrix metalloproteinase-1 expression and cell invasion in Helicobacter pylori infection. Gut 2013; 62:358–367.
Zhang W, Chen DQ, Qi F, Wang J, Xiao WY, Zhu WZ . Inhibition of calcium-calmodulin-dependent kinase II suppresses cardiac fibroblast proliferation and extracellular matrix secretion. J Cardiovasc Pharmacol 2010; 55:96–105.
Kim HH, Shin CM, Park CH, et al. Eicosapentaenoic acid inhibits UV-induced MMP-1 expression in human dermal fibroblasts. J Lipid Res 2005; 46:1712–1720.
Reunanen N, Li SP, Ahonen M, Foschi M, Han J, Kahari VM . Activation of p38α MAPK enhances collagenase-1 (matrix metalloproteinase (MMP)-1) and stromelysin-1 (MMP-3) expression by mRNA stabilization. J Biol Chem 2002; 277:32360–32368.
Korzus E, Nagase H, Rydell R, Travis J . The mitogen-activated protein kinase and JAK-STAT signaling pathways are required for an oncostatin M-responsive element-mediated activation of matrix metalloproteinase 1 gene expression. J Biol Chem 1997; 272:1188–1196.
Sebastian C, Zwaans BM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012; 151:1185–1199.
Zhang ZG, Qin CY . Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signal-regulated kinase signaling pathway. Mol Med Rep 2014; 9:882–888.
Kugel S, Feldman JL, Klein MA, et al. Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer. Cell Rep 2015; 13:479–488.
Sundaresan NR, Vasudevan P, Zhong L, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med 2012; 18:1643–1650.
Mertens AE, Roovers RC, Collard JG . Regulation of Tiam1-Rac signalling. FEBS Lett 2003; 546:11–16.
Eblen ST, Slack JK, Weber MJ, Catling AD . Rac-PAK signaling stimulates extracellular signal-regulated kinase (ERK) activation by regulating formation of MEK1-ERK complexes. Mol Cell Biol 2002; 22:6023–6033.
Soto E, Yanagisawa M, Marlow LA, Copland JA, Perez EA, Anastasiadis PZ . p120 catenin induces opposing effects on tumor cell growth depending on E-cadherin expression. J Cell Biol 2008; 183:737–749.
Vougiouklakis T, Sone K, Saloura V, et al. SUV420H1 enhances the phosphorylation and transcription of ERK1 in cancer cells. Oncotarget 2015; 6:43162–43171.
Hunker CM, Giambini H, Galvis A, et al. Rin1 regulates insulin receptor signal transduction pathways. Exp Cell Res 2006; 312:1106–1118.
Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res 2014; 20:4107–4114.
Sparano JA, Bernardo P, Stephenson P, et al. Randomized phase III trial of marimastat versus placebo in patients with metastatic breast cancer who have responding or stable disease after first-line chemotherapy: Eastern Cooperative Oncology Group trial E2196. J Clin Oncol 2004; 22:4683–4690.
Yeh TC, Marsh V, Bernat BA, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res 2007; 13:1576–1583.
Wisniewski JR, Zougman A, Nagaraj N, Mann M . Universal sample preparation method for proteome analysis. Nat Methods 2009; 6:359–362.
Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007; 448:807–810.
Acknowledgements
We thank Drs Lei Zhang, Xiumin Yan, Zhuang Wei, Shuai Wu, Lin Pan and Liang Dong for technical supports. We thank Dangsheng Li for the critical reading of the manuscript and suggestions. We also thank all of our lab members for helpful discussion. This work was supported by the Ministry of Science and Technology of China (2012CB910800), the National Natural Science Foundation of China (81430066, 81402276, 81402371, 81401898, 81402498, 81101583, 81372509, 31370747 and 81325015), the Science and Technology Commission of Shanghai Municipality (12JC1409800, 15XD1504000), the “Cross and cooperation in science and technology innovation team” program, the Chinese Postdoctoral foundation (2014M561536, 2013T60476), the Shanghai Postdoctoral Foundation (14R21411400), and the Shanghai Institutes for Biological Sciences (2013KIP303, 2013KIP102, 2014KIP304).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
409 potential gene fusions or isoforms in ADCs. (XLS 53 kb)
Supplementary information, Table S2
284 potential gene fusions or isoforms in SCCs. (XLS 43 kb)
Supplementary information, Table S3
Clinical features of 5 lung SCC patients harboring TRA2B-DNAH5 fusion (PDF 8 kb)
Supplementary information, Table S4
Identification of TRA2B-DNAH5 fusion interacting proteins IP-MS analyses. (XLS 24 kb)
Supplementary information, Table S5
The list of primers used for real-time PCR quantification in this study. (PDF 19 kb)
Supplementary information, Figure S1
Identification of TIE1 fusion in human lung ADC. (PDF 172 kb)
Supplementary information, Figure S2
Identification of genomic breakpoint of TRA2B-DNAH5 fusion. (PDF 153 kb)
Supplementary information, Figure S3
Sanger sequencing confirms the TRA2B-DNAH5 fusion in another 4 human lung SCC specimensat RNA level. (PDF 122 kb)
Supplementary information, Figure S4
The TRA2B-DNAH5 fusion promotes cell transformation. (PDF 51 kb)
Supplementary information, Figure S5
Immunohistochemical studies of CRL-5889 xenograft tumors. (PDF 154 kb)
Supplementary information, Figure S6
Confirmation of MMP1 up-regulation and ERK1/2 activation in cells and xenograft tumors. (PDF 166 kb)
Supplementary information, Figure S7
Up-regulated MMP1 expression mediates the promotive function of TRA2B-DNAH5 fusion in cell invasion and migration. (PDF 142 kb)
Supplementary information, Figure S8
Knockdown of MMP1 significantly inhibits the tumor-promotivefunction of the TRA2B-DNAH5 fusion in CRL-5889 xenograft assay. (PDF 114 kb)
Supplementary information, Figure S9
Human lung SCC specimens harboring the TRA2B-DNAH5 fusion are stained positive for p-ERK1/2 and MMP1. (PDF 172 kb)
Supplementary information, Figure S10
TRA2B-DNAH5 fusion is mainlylocated at cytoplasm. (PDF 52 kb)
Supplementary information, Figure S11
CHIP-Seq data reveals the binding of SIRT6 on TIAM1, SUV620H1 and RIN1 genes. (PDF 71 kb)
Supplementary information, Figure S12
IGF1R may be involved in the activation of MEK1/2 and up-regulation of MMP1 by TRA2B-DNAH5 fusion. (PDF 75 kb)
Supplementary information, Figure S13
Human lung SCC cells expressing the TRA2B-DNAH5 fusion are more sensitive to selumetinib treatment. (PDF 118 kb)
Supplementary information, Figure S14
Selumetinib treatment inhibits ERK1/2 activity and MMP1 levels in CRL-5889 xenograft tumors with TRA2B-DNAH5 fusion expression. (PDF 164 kb)
Supplementary information, Figure S15
Selumetinib treatment doesn't cause a significant weight loss in mice. (PDF 112 kb)
Supplementary information, Figure S16
Marimastat treatment has no inhibitory effect on tumor growth of CRL-5889 xenograft tumors. (PDF 61 kb)
Supplementary information, Data S1
CDS sequence and amino acids sequence of TRA2B-DNAH5 fusion. (PDF 44 kb)
Rights and permissions
About this article
Cite this article
Li, F., Fang, Z., Zhang, J. et al. Identification of TRA2B-DNAH5 fusion as a novel oncogenic driver in human lung squamous cell carcinoma. Cell Res 26, 1149–1164 (2016). https://doi.org/10.1038/cr.2016.111
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2016.111
Keywords
This article is cited by
-
Switched alternative splicing events as attractive features in lung squamous cell carcinoma
Cancer Cell International (2022)
-
Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression
Scientific Reports (2021)
-
Identification of a novel gene signature for the prediction of recurrence in HCC patients by machine learning of genome-wide databases
Scientific Reports (2020)