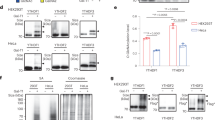
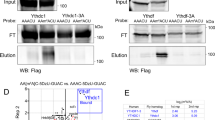
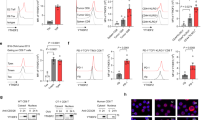
Abstract
N6-methyladenosine (m6A) is the most abundant internal modification in eukaryotic messenger RNAs (mRNAs), and plays important roles in cell differentiation and tissue development. It regulates multiple steps throughout the RNA life cycle including RNA processing, translation, and decay, via the recognition by selective binding proteins. In the cytoplasm, m6A binding protein YTHDF1 facilitates translation of m6A-modified mRNAs, and YTHDF2 accelerates the decay of m6A-modified transcripts. The biological function of YTHDF3, another cytoplasmic m6A binder of the YTH (YT521-B homology) domain family, remains unknown. Here, we report that YTHDF3 promotes protein synthesis in synergy with YTHDF1, and affects methylated mRNA decay mediated through YTHDF2. Cells deficient in all three YTHDF proteins experience the most dramatic accumulation of m6A-modified transcripts. These results indicate that together with YTHDF1 and YTHDF2, YTHDF3 plays critical roles to accelerate metabolism of m6A-modified mRNAs in the cytoplasm. All three YTHDF proteins may act in an integrated and cooperative manner to impact fundamental biological processes related to m6A RNA methylation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Desrosiers R, Friderici K, Rottman F . Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 1974; 71:3971–3975.
Wei C, Gershowitz A, Moss B . 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 1976; 15:397–401.
Wei C, Moss B . Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 1977; 16:1672–1676.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201–206.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR . Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635–1646.
Batista PJ, Molinie B, Wang J, et al. M6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014; 15:707–719.
Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. M6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015; 347:1002–1006.
Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793–806.
Fischer J, Koch L, Emmerling C, et al. Inactivation of the Fto gene protects from obesity. Nature 2009; 458:894–898.
Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014; 24:1403–1419.
Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013; 49:18–29.
Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505:117–120.
Wang X, Zhao BS, Roundtree IA, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015; 161:1388–1399.
Xu C, Wang X, Liu K, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 2014; 10:927–929.
Xiao W, Adhikari S, Dahal U, et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 2016; 61:507–519.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF . HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 2015; 162:1299–1308.
Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T . N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015; 518:560–564.
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian S-B . Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015; 526:591–594.
Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014; 10:93–95.
Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E . mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 2006; 20:1885–1898.
Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN . Y-box-binding protein 1 and its functions. Biochemistry 2011; 76:1402–1433.
Le Hir H, Séraphin B . EJCs at the heart of translational control. Cell 2008; 133:213–216.
Li F, Zhao D, Wu J, Shi Y . Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res 2014; 24:1490–1492.
Zhu T, Roundtree IA, Wang P, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res 2014; 24:1493–1496.
Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J . Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem 2015; 290:24902–24913.
Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6:pl1.
Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401–404.
Lu Y, He X, Zhong S . Cross-species microarray analysis with the OSCAR system suggests an INSR->Pax6->NQO1 neuro-protective pathway in aging and Alzheimer's disease. Nucleic Acids Res 2007; 35:105–114.
Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753–767.
Fritzsche R, Karra D, Bennett KL, et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep 2013; 5:1749–1762.
Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007; 445:168–176.
Medioni C, Mowry K, Besse F . Principles and roles of mRNA localization in animal development. Development 2012; 139:3263–3276.
Birkaya B, Ortt K, Sinha S . Novel in vivo targets of ΔNp63 in keratinocytes identified by a modified chromatin immunoprecipitation approach. BMC Mol Biol 2007; 8:43.
Aguilo F, Zhang F, Sancho A, et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 2015; 17:689–704.
Nagaraja Tirumuru, Boxuan Simen Zhao, Wuxun Lu, Zhike Lu CH, Li W . N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 2016; 5:e15528
Kennedy EM, Bogerd HP, Kornepati AVR, et al. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 2016; 19:675–685.
Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM . Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437–445.
Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE . Y14 and hUpf3b form an NMD-activating complex. Mol Cell 2003; 11:939–949.
Guet CC, Bruneaux L, Min TL, et al. Minimally invasive determination of mRNA concentration in single living bacteria. Nucleic Acids Res 2008; 36:e73.
Jia G, Fu Y, Zhao X, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885–887.
Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS . Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009; 324:218–223.
Gandin V, Sikström K, Alain T, et al. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp 2014; 87:1–10.
Pearson WR, Wood T, Zhang Z, Miller W . Comparison of DNA sequences with protein sequences. Genomics 1997; 46:24–36.
Trapnell C, Pachter L, Salzberg SL . TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25:1105–1111.
Anders S, Huber W . Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106.
Corcoran DL, Georgiev S, Mukherjee N, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 2011; 12:R79.
Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38:576–589.
Bazzini AA, Lee MT, Giraldez AJ . Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012; 336:233–237.
Huang DW, Lempicki RA, Sherman BT . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57.
Huang DW, Sherman BT, Lempicki RA . Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1–13.
Fran Supek, Matko Bošnjak, Nives Škunca TŠ . Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One 2011; 6: e21800.
Koressaar T, Remm M . Enhancements and modifications of primer design program Primer3. Bioinformatics 2007; 23:1289–1291.
Untergasser A, Cutcutache I, Koressaar T, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res 2012; 40: e115.
Shi Y, Sawada J, Sui G, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003; 422:735–738.
Acknowledgements
We thank Professor T Pan (The University of Chicago) for kindly providing the polysome profiling instrument. We thank Dr J Tauler (The University of Chicago) for editing the manuscript. We also thank Institutes of Biomedical Sciences at Fudan University for the protein mass spectrometry experiments. The work was supported by the National Institutes of Health grants GM071440 and GM113194 to CH. CH is an investigator of the Howard Hughes Medical Institute (HHMI). HS is supported by the Gustavus F Swift Fellowship from the Chemistry Department at University of Chicago. HM is supported by the Postdoctoral International Exchange Program of the China Postdoctoral Council (CPC). The Mass Spectrometry Facility of the University of Chicago is funded by the National Science Foundation (CHE-1048528).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Analyses of YTHDF3 PAR-CLIP data. (PDF 271 kb)
Supplementary information, Figure S2
Ribosome profiling and polysome profiling of HeLa cells depleted of YTHDF3 (A-J), and protein and mRNA levels of reporters in the double tethering assay (K). (PDF 210 kb)
Supplementary information, Figure S3
YTHDF3 and YTHDF1 co-immunoprecipitate and share protein partners. (PDF 103 kb)
Supplementary information, Figure S4
YTHDF3 regulates binding specificity and affinity of THDF1 and YTHDF2 towards target RNAs. (PDF 114 kb)
Supplementary information, Figure S5
Temporal order of YTHDFs' interaction with m6A-modified transcripts. (PDF 126 kb)
Supplementary information, Table S1
Target transcripts of YTHDF3 and Comparison of YTHDF1-3 targets (XLSX 25543 kb)
Supplementary information, Table S2
YTHDF3 protein interactome and Comparison of YTHDF1 and YTHDF3 protein partners (XLSX 39 kb)
Rights and permissions
About this article
Cite this article
Shi, H., Wang, X., Lu, Z. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 27, 315–328 (2017). https://doi.org/10.1038/cr.2017.15
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2017.15
Keywords
This article is cited by
-
RNA epigenetic modifications as dynamic biomarkers in cancer: from mechanisms to clinical translation
Biomarker Research (2025)
-
Targeting m6A methylation for early diagnosis and precision medicine in hepatocellular carcinoma
Cancer Cell International (2025)
-
Fto-mediated m6A modification is essential for cerebellar development through regulating epigenetic reprogramming
Journal of Biomedical Science (2025)
-
Biological roles of enhancer RNA m6A modification and its implications in cancer
Cell Communication and Signaling (2025)
-
From bone marrow mesenchymal stem cells to diseases: the crucial role of m6A methylation in orthopedics
Stem Cell Research & Therapy (2025)