Abstract
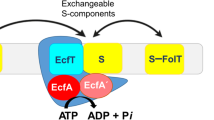
Energy-coupling factor (ECF) transporters are a large family of ATP-binding cassette transporters recently identified in microorganisms. Responsible for micronutrient uptake from the environment, ECF transporters are modular transporters composed of a membrane substrate-binding component EcfS and an ECF module consisting of an integral membrane scaffold component EcfT and two cytoplasmic ATP binding/hydrolysis components EcfA/A'. ECF transporters are classified into groups I and II. Currently, the molecular understanding of group-I ECF transporters is very limited, partly due to a lack of transporter complex structural information. Here, we present structures and structure-based analyses of the group-I cobalt ECF transporter CbiMNQO, whose constituting subunits CbiM/CbiN, CbiQ, and CbiO correspond to the EcfS, EcfT, and EcfA components of group-II ECF transporters, respectively. Through reconstitution of different CbiMNQO subunits and determination of related ATPase and transporter activities, the substrate-binding subunit CbiM was found to stimulate CbiQO's basal ATPase activity. The structure of CbiMQO complex was determined in its inward-open conformation and that of CbiO in β, γ-methyleneadenosine 5′-triphosphate-bound closed conformation. Structure-based analyses revealed interactions between different components, substrate-gating function of the L1 loop of CbiM, and conformational changes of CbiO induced by ATP binding and product release within the CbiMNQO transporter complex. These findings enabled us to propose a working model of the CbiMNQO transporter, in which the transport process requires the rotation or toppling of both CbiQ and CbiM, and CbiN might function in coupling conformational changes between CbiQ and CbiM.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Locher KP, Lee AT, Rees DC . The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 2002; 296:1091–1098.
Hvorup RN, Goetz BA, Niederer M, Hollenstein K, Perozo E, Locher KP . Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science 2007; 317:1387–1390.
Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J . Crystal structure of a catalytic intermediate of the maltose transporter. Nature 2007; 450:515–521.
Oldham ML, Chen J . Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 2011; 332:1202–1205.
Khare D, Oldham ML, Orelle C, Davidson AL, Chen J . Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell 2009; 33:528–536.
Hollenstein K, Frei DC, Locher KP . Structure of an ABC transporter in complex with its binding protein. Nature 2007; 446:213–216.
Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC . The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 2008; 321:250–253.
Gerber S, Comellas-Bigler M, Goetz BA, Locher KP . Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 2008; 321:246–250.
Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T . Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J Bacteriol 2006; 188:317–327.
Duurkens RH, Tol MB, Geertsma ER, Permentier HP, Slotboom DJ . Flavin binding to the high affinity riboflavin transporter RibU. J Biol Chem 2007; 282:10380–10386.
Hebbeln P, Rodionov DA, Alfandega A, Eitinger T . Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci USA 2007; 104:2909–2914.
Rodionov DA, Hebbeln P, Eudes A, et al. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 2009; 191:42–51.
Rees DC, Johnson E, Lewinson O . ABC transporters: the power to change. Nat Rev Mol Cell Biol 2009; 10:218–227.
Chen J . Molecular mechanism of the Escherichia coli maltose transporter. Curr Opin Struct Biol 2013; 23:492–498.
Zolnerciks JK, Andress EJ, Nicolaou M, Linton KJ . Structure of ABC transporters. Essays Biochem 2011; 50:43–61.
Locher KP . Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol 2016; 23:487–493.
Zhang P, Wang J, Shi Y . Structure and mechanism of the S component of a bacterial ECF transporter. Nature 2010; 468:717–720.
Erkens GB, Berntsson RP, Fulyani F, et al. The structural basis of modularity in ECF-type ABC transporters. Nat Struct Mol Biol 2011; 18:755–760.
Berntsson RP, ter Beek J, Majsnerowska M, et al. Structural divergence of paralogous S components from ECF-type ABC transporters. Proc Natl Acad Sci USA 2012; 109:13990–13995.
Zhao Q, Wang C, Wang C, et al. Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters. Nat Commun 2015; 6:7661.
Eudes A, Erkens GB, Slotboom DJ, Rodionov DA, Naponelli V, Hanson AD . Identification of genes encoding the folate- and thiamine-binding membrane proteins in Firmicutes. J Bacteriol 2008; 190:7591–7594.
Zhang P . Structure and mechanism of energy-coupling factor transporters. Trends Microbiol 2013; 21:652–659.
Wang T, Fu G, Pan X, et al. Structure of a bacterial energy-coupling factor transporter. Nature 2013; 497:272–276.
Xu K, Zhang M, Zhao Q, et al. Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature 2013; 497:268–271.
Zhang M, Bao Z, Zhao Q, et al. Structure of a pantothenate transporter and implications for ECF module sharing and energy coupling of group II ECF transporters. Proc Natl Acad Sci USA 2014; 111:18560–18565.
Slotboom DJ . Structural and mechanistic insights into prokaryotic energy-coupling factor transporters. Nat Rev Microbiol 2014; 12:79–87.
Finkenwirth F, Sippach M, Landmesser H, et al. ATP-dependent conformational changes trigger substrate capture and release by an ECF-type biotin transporter. J Biol Chem 2015; 290:16929–16942.
Okamoto S, Eltis LD . The biological occurrence and trafficking of cobalt. Metallomics 2011; 3:963–970.
Lebrette H, Brochier-Armanet C, Zambelli B, et al. Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins. Structure 2014; 22:1421–1432.
Desguin B, Goffin P, Viaene E, et al. Lactate racemase is a nickel-dependent enzyme activated by a widespread maturation system. Nat Commun 2014; 5:3615.
Yu Y, Zhou M, Kirsch F, et al. Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters. Cell Res 2013; 24:267–277.
Siche S, Neubauer O, Hebbeln P, Eitinger T . A bipartite S unit of an ECF-type cobalt transporter. Res Microbiol 2010; 161:824–829.
Kirsch F, Eitinger T . Transport of nickel and cobalt ions into bacterial cells by S components of ECF transporters. Biometals 2014; 27:653–660.
Neubauer O, Alfandega A, Schoknecht J, Sternberg U, Pohlmann A, Eitinger T . Two essential arginine residues in the T components of energy-coupling factor transporters. J Bacteriol 2009; 191:6482–6488.
Orelle C, Ayvaz T, Everly RM, Klug CS, Davidson AL . Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc Natl Acad Sci USA 2008; 105:12837–12842.
Davidson AL, Shuman HA, Nikaido H . Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci USA 1992; 89:2360–2364.
Liu CE, Liu PQ, Ames GF . Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter). J Biol Chem 1997; 272:21883–21891.
Qi X, Lin W, Ma M, et al. Structural basis of rifampin inactivation by rifampin phosphotransferase. Proc Natl Acad Sci USA 2016; 113:3803–3808.
Minor W, Cymborowski M, Otwinowski Z, Chruszcz M . HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 2006; 62:859–866.
Kabsch W . Xds. Acta Crystallogr D Biol Crystallogr 2010; 66:125–132.
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ . Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674.
Cowtan K . Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr 2010; 66:470–478.
Emsley P, Cowtan K . Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132.
Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213–221.
Nielsen M, Lundegaard C, Lund O, Petersen TN . CPHmodels-3.0--remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res 2010; 38:W576–581.
Tang WK, Xia D . Role of the D1-D2 linker of human VCP/p97 in the asymmetry and ATPase activity of the D1-domain. Sci Rep 2016; 6:20037.
Acknowledgements
We thank the staff members at BL19U/18U of National Center for Protein Science Shanghai (NCPSS) and BL17U of Shanghai Synchrotron Radiation Facility (SSRF) for technical assistance in data collection, and the staff at the core facility center of Institute of Plant Physiology and Ecology for X-ray diffraction experimental test and analysis. We thank Dr David Degen from Rutgers University for manuscript reading and revision. This work was supported by grants from the Ministry of Science and Technology of China (2015CB910900 to PZ and 2015CB910104 to JW), the National Natural Science Foundation of China (31670755 and 31322016 to PZ), the Chinese Academy of Sciences (QYZDB-SSW-SMC006 to PZ and 2015LH0027 to FH), and the Shanghai Municipal Science and Technology Commission. Research in DL Lab is supported by the 1000 Young Talent Program, the Shanghai “Pujiang Talent” Program (15PJ1409400), and the National Natural Science Foundation of China (31570748).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
SDS-PAGE and western blot results of the samples during the purification process. (PDF 275 kb)
Supplementary information, Figure S2
Representative 2FoFc electron density contoured at 1.5σ level. (PDF 307 kb)
Supplementary information, Figure S3
Residue Phe75 from TM4 docks its side chain into the empty substrate binding site of CbiM (PDF 121 kb)
Supplementary information, Figure S4
Sequence alignment of CbiQ, NikQ and group-II EcfT from different species. (PDF 464 kb)
Supplementary information, Figure S5
Hydrogen-bonding Interactions between CH2-CH3 of CbiQ and CbiO. (PDF 448 kb)
Supplementary information, Figure S6
Sequence alignment of CbiM and NikM. (PDF 477 kb)
Supplementary information, Figure S7
A model shows the uncoupling of ATPase activity with cobalt transport caused by loss of CbiN. (PDF 113 kb)
Supplementary information, Table S1
Statistics of data collection and structure refinement. (PDF 108 kb)
Rights and permissions
About this article
Cite this article
Bao, Z., Qi, X., Hong, S. et al. Structure and mechanism of a group-I cobalt energy coupling factor transporter. Cell Res 27, 675–687 (2017). https://doi.org/10.1038/cr.2017.38
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2017.38
Keywords
This article is cited by
-
Single-molecule visualization of ATP-induced dynamics of the subunit composition of an ECF transporter complex under turnover conditions
Nature Communications (2025)
-
Cryo-EM structure and molecular mechanism of the jasmonic acid transporter ABCG16
Nature Plants (2024)
-
Cryo-EM structure and molecular mechanism of abscisic acid transporter ABCG25
Nature Plants (2023)
-
Identification of inhibitors targeting the energy-coupling factor (ECF) transporters
Communications Biology (2023)
-
Expulsion mechanism of the substrate-translocating subunit in ECF transporters
Nature Communications (2023)