Abstract
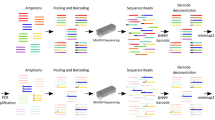
Resequencing of genomic regions that have been implicated by linkage and/or association studies to harbor genetic susceptibility loci represents a necessary step to identify causal variants. Massively parallel sequencing (MPS) offers the possibility of SNP discovery and frequency determination among pooled DNA samples. The strategies of pooling DNA samples and pooling PCR amplicons generated from individual DNA samples were evaluated, and both were found to return accurate estimates of SNP frequencies across varying levels of sequence coverage.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Margulies M, Egholm M, Altman WE et al: Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437: 376–380.
Engelmark MT, Ivansson EL, Magnusson JJ et al: Identification of susceptibility loci for cervical carcinoma by genome scan of affected sib-pairs. Hum Mol Genet 2006; 15: 3351–3360.
Sherry ST, Ward MH, Kholodov M et al: dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–311.
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM : Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 2007; 8: R143.
Albert TJ, Molla MN, Muzny DM et al: Direct selection of human genomic loci by microarray hybridization. Nat Methods 2007; 4: 903–905.
Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M : Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res 2005; 33: e71.
Hodges E, Xuan Z, Balija V et al: Genome-wide in situ exon capture for selective resequencing. Nat Genet 2008; 39: 1522–1527.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Ingman, M., Gyllensten, U. SNP frequency estimation using massively parallel sequencing of pooled DNA. Eur J Hum Genet 17, 383–386 (2009). https://doi.org/10.1038/ejhg.2008.182
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2008.182
Keywords
This article is cited by
-
SNP allele frequency estimation in pooled DNA of Frankliniella occidentalis (Pergande, 1895) (Thysanoptera: Thripidae) by next-generation sequencing
Journal of Plant Diseases and Protection (2023)
-
Detection and mapping of mtDNA SNPs in Atlantic salmon using high throughput DNA sequencing
BMC Genomics (2011)
-
Estimation of allele frequency and association mapping using next-generation sequencing data
BMC Bioinformatics (2011)
-
SNP identification, verification, and utility for population genetics in a non-model genus
BMC Genetics (2010)