Abstract
Mutations that affect splicing of precursor messenger RNAs play a major role in the development of hereditary diseases. Most splicing mutations have been found to eliminate GT or AG dinucleotides that define the 5′ and 3′ ends of introns, leading to exon skipping or cryptic splice-site activation. Although accurate description of the mis-spliced transcripts is critical for predicting phenotypic consequences of these alterations, their exact nature in affected individuals cannot often be determined experimentally. Using a comprehensive collection of exons that sustained cryptic splice-site activation or were skipped as a result of splice-site mutations, we have developed a multivariate logistic discrimination procedure that distinguishes the two aberrant splicing outcomes from DNA sequences. The new algorithm was validated using an independent sample of exons and implemented as a free online utility termed CRYP-SKIP (http://www.dbass.org.uk/cryp-skip/). The web application takes up one or more mutated alleles, each consisting of one exon and flanking intronic sequences, and provides a list of important predictor variables and their values, the overall probability of activating cryptic splice vs exon skipping, and the location and intrinsic strength of predicted cryptic splice sites in the input sequence. These results will facilitate phenotypic prediction of splicing mutations and provide further insights into splicing enhancer and silencer elements and their relative importance for splice-site selection in vivo.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Buratti E, Baralle FE : Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 2004; 24: 10505–10514.
Hiller M, Zhang Z, Backofen R, Stamm S : Pre-mRNA secondary structures influence exon recognition. PLoS Genet 2007; 3: e204.
Liu HX, Zhang M, Krainer AR : Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev 1998; 12: 1998–2012.
Graveley BR, Hertel KJ, Maniatis T : SR proteins are ‘locators’ of the RNA splicing machinery. Curr Biol 1999; 9: R6–R7.
Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G : hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res 1992; 20: 3671–3678.
Matunis MJ, Xing J, Dreyfuss G : The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res 1994; 22: 1059–1067.
Burd CG, Dreyfuss G : RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J 1994; 13: 1197–1204.
Caputi M, Zahler AM : Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J Biol Chem 2001; 276: 43850–43859.
Schaal TD, Maniatis T : Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol 1999; 19: 1705–1719.
Singh NN, Androphy EJ, Singh RN : In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA 2004; 10: 1291–1305.
Fairbrother WG, Yeh RF, Sharp PA, Burge CB : Predictive identification of exonic splicing enhancers in human genes. Science 2002; 297: 1007–1013.
Zhang XH, Chasin LA : Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 2004; 18: 1241–1250.
Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB : Systematic identification and analysis of exonic splicing silencers. Cell 2004; 119: 831–845.
Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA : Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol 2005; 25: 7323–7332.
Goren A, Ram O, Amit M et al: Comparative analysis identifies exonic splicing regulatory sequences–the complex definition of enhancers and silencers. Mol Cell 2006; 22: 769–781.
Wimmer K, Roca X, Beiglbock H et al: Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum Mutat 2007; 28: 599–612.
Kralovicova J, Vorechovsky I : Global control of aberrant splice site activation by auxiliary splicing sequences: evidence for a gradient in exon and intron definition. Nucleic Acids Res 2007; 35: 6399–6413.
Wang Z, Xiao X, Van Nostrand E, Burge CB : General and specific functions of exonic splicing silencers in splicing control. Mol Cell 2006; 23: 61–70.
Ars E, Serra E, Garcia J et al: Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet 2000; 9: 237–247.
Teraoka SN, Telatar M, Becker-Catania S et al: Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet 1999; 64: 1617–1631.
Cooper DN, Krawczak M : Human Gene Mutation. Oxford: BIOS Scientific Publishers, 1993.
Shapiro MB, Senapathy P : RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 1987; 15: 7155–7174.
Vorechovsky I : Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res 2006; 34: 4630–4641.
Buratti E, Chivers MC, Kralovicova J et al: Aberrant 5′ splice sites in human disease genes: mutation pattern, nucletide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res 2007; 35: 4250–4263.
Reese MG, Eeckman FH, Kulp D, Haussler D : Improved splice site detection in Genie. J Comput Biol 1997; 4: 311–323.
Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR : An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 2006; 15: 2490–2508.
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR : ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003; 31: 3568–3571.
Zhang C, Li WH, Krainer AR, Zhang MQ : RNA landscape of evolution for optimal exon and intron discrimination. Proc Natl Acad Sci USA 2008; 105: 5797–5802.
Stadler MB, Shomron N, Yeo GW, Schneider A, Xiao X, Burge CB : Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet 2006; 2: e191.
Team RDC : R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing: ISBN 3-900051-07-0 2007.
Houdayer C, Dehainault C, Mattler C et al: Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum Mutat 2008; 29: 975–982.
Carmel I, Tal S, Vig I, Ast G : Comparative analysis detects dependencies among the 5′ splice-site positions. RNA 2004; 10: 828–840.
Pan Q, Bakowski MA, Morris Q et al: Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet 2005; 21: 73–77.
Meili D, Kralovicova J, Zagalak J et al: Disease-causing mutations improving the branch site and polypyrimidine tract: pseudoexon activation of LINE-2 and antisense Alu lacking the poly(T)-tail. Hum Mutat 2009; (in press).
Reed R : The organization of 3′ splice-site sequences in mammalian introns. Genes Dev 1989; 3: 2113–2123.
McCullough AJ, Berget SM : G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol 1997; 17: 4562–4571.
Han K, Yeo G, An P, Burge CB, Grabowski PJ : A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol 2005; 3: e158.
Kralovicova J, Vorechovsky I : Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J Immunol 2006; 176: 2381–2388.
Acknowledgements
We thank Sarah Ennis and Nicholas Maniatis for useful discussions and critical reading of the manuscript. This work was funded by a Grant from the JDRF (2008-47) to IV.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Divina, P., Kvitkovicova, A., Buratti, E. et al. Ab initio prediction of mutation-induced cryptic splice-site activation and exon skipping. Eur J Hum Genet 17, 759–765 (2009). https://doi.org/10.1038/ejhg.2008.257
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2008.257
Keywords
This article is cited by
-
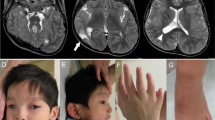
Exome sequencing implicates genetic disruption of prenatal neuro-gliogenesis in sporadic congenital hydrocephalus
Nature Medicine (2020)
-
A novel splicing mutation in SLC9A6 in a boy with Christianson syndrome
Human Genome Variation (2019)
-
Splicing mutations in human genetic disorders: examples, detection, and confirmation
Journal of Applied Genetics (2018)
-
IntSplice: prediction of the splicing consequences of intronic single-nucleotide variations in the human genome
Journal of Human Genetics (2016)
-
An ENU mutagenesis screen identifies novel and known genes involved in epigenetic processes in the mouse
Genome Biology (2013)