Abstract
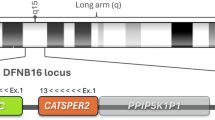
In communities with high rates of consanguinity and consequently high prevalence of recessive phenotypes, homozygosity mapping with SNP arrays is an effective approach for gene discovery. In 20 Palestinian kindreds with prelingual nonsyndromic hearing loss, we generated homozygosity profiles reflecting linkage to the phenotype. Family sizes ranged from small nuclear families with two affected children, one unaffected sibling, and parents to multigenerational kindreds with 12 affected relatives. By including unaffected parents and siblings and screening 250 K SNP arrays, even small nuclear families yielded informative profiles. In 14 families, we identified the allele responsible for hearing loss by screening a single candidate gene in the longest homozygous region. Novel alleles included missense, nonsense, and splice site mutations of CDH23, MYO7A, MYO15A, OTOF, PJVK, Pendrin/SLC26A4, TECTA, TMHS, and TMPRSS3, and a large genomic deletion of Otoancorin (OTOA). All point mutations were rare in the Palestinian population (zero carriers in 288 unrelated controls); the carrier frequency of the OTOA genomic deletion was 1%. In six families, we identified five genomic regions likely to harbor novel genes for human hearing loss on chromosomes 1p13.3 (DFNB82), 9p23–p21.2/p13.3–q21.13 (DFNB83), 12q14.3–q21.2 (DFNB84; two families), 14q23.1–q31.1, and 17p12–q11.2 (DFNB85).
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Jaber L, Halpern GJ, Shohat T : Trends in the frequencies of consanguineous marriages in the Israeli Arab community. Clin Genet 2000; 58: 106–110.
Zlotogora J, Barges S, Bisharat B, Shalev SA : Genetic disorders among Palestinian Arabs. 4: Genetic clinics in the community. Am J Med Genet A 2006; 140: 1644–1646.
Yasunaga S, Grati M, Cohen-Salmon M et al: A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet 1999; 21: 363–369.
Mustapha M, Weil D, Chardenoux S et al: An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet 1999; 8: 409–412.
Zwaenepoel I, Mustapha M, Leibovici M et al: Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci USA 2002; 99: 6240–6245.
Scott HS, Kudoh J, Wattenhofer M et al: Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet 2001; 27: 59–63.
Mburu P, Mustapha M, Varela A et al: Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet 2003; 34: 421–428.
Shahin H, Walsh T, Sobe T et al: Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive non-syndromic hearing loss. Am J Hum Genet 2006; 78: 144–152.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Shahin H, Walsh T, Sobe T et al: Genetics of congenital deafness in the Palestinian population: multiple connexin 26 alleles with shared origins in the Middle East. Hum Genet 2002; 110: 284–289.
Walsh T, Abu Rayan A, Abu Sa'ed J et al: Genomic analysis of a heterogeneous Mendelian phenotype: multiple novel alleles for inherited hearing loss in the Palestinian population. Hum Genomics 2006; 2: 203–211.
Ott J : Analysis of Human Genetic Linkage, 3rd edn., Johns Hopkins Univ Press: Baltimore.
Hereditary Hearing Loss Homepage. ( http://webh01.ua.ac.be/hhh/ ).
Selzer RR, Richmond TA, Pofahl NJ et al: Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 2005; 44: 305–319.
Livak KJ, Schmittgen TD : Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Methods. Methods 2001; 25: 402–408.
Itsara A, Cooper GM, Baker C et al: Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 2009; 84: 148–161.
Masmoudi S, Tlili A, Majava M et al: Mapping of a new autosomal recessive nonsyndromic hearing loss locus (DFNB32) to chromosome 1p13.3–22 1. Eur J Hum Genet 2003; 11: 185–188.
Kurima K, Peters LM, Yang Y et al: Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet 2002; 30: 277–284.
Williamson RE, Darrow KN, Michaud S et al: Methylthioadenosine phosphorylase (MTAP) in hearing: Gene disruption by chromosomal rearrangement in a hearing impaired individual and model organism analysis. Am J Med Genet A 2007; 143A: 1630–1639.
D'Adamo P, Donaudy F, D'Eustacchio A, Di Iorio E, Melchionda S, Gasparini P : A new locus (DFNA47) for autosomal dominant non-syndromic inherited hearing loss maps to 9p21–22 in a large Italian family. Eur J Hum Genet 2003; 11: 121–124.
Collin RW, Kalay E, Tariq M et al: Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am J Hum Genet 2008; 82: 125–138.
Zhang Y, Malekpour M, Al-Madani N et al: Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J Med Genet 2007; 44: 233–240.
Knijnenburg J, Oberstein SA, Frei K et al: A homozygous deletion of a normal variation locus in a patient with hearing loss from non-consanguineous parents. J Med Genet 2009; 46: 412–417.
Acknowledgements
We thank the families for their generous participation in this study. We thank Dr Wael Salhab for a clinical evaluation of the patients, and Michael Rahil of the Dar Al-Kalima Audiology Clinic for audiological analyses. We thank the technical staff of the Hereditary Research Laboratory (Bethlehem University), Rob Hall at the Center for Array Technology (University of Washington), Gregory Cooper, and Heather Mefford for helpful comments and suggestions. This study was supported by the NIH/NIDCD Grant R01DC005641.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Shahin, H., Walsh, T., Rayyan, A. et al. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur J Hum Genet 18, 407–413 (2010). https://doi.org/10.1038/ejhg.2009.190
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2009.190
Keywords
This article is cited by
-
The natural history, clinical outcomes, and genotype–phenotype relationship of otoferlin-related hearing loss: a systematic, quantitative literature review
Human Genetics (2023)
-
Analysis of the genotype–phenotype correlation of MYO15A variants in Chinese non-syndromic hearing loss patients
BMC Medical Genomics (2022)
-
ARNSHL gene identification: past, present and future
Molecular Genetics and Genomics (2022)
-
Identification of autosomal recessive nonsyndromic hearing impairment genes through the study of consanguineous and non-consanguineous families: past, present, and future
Human Genetics (2022)
-
Comprehensive genetic testing of Chinese SNHL patients and variants interpretation using ACMG guidelines and ethnically matched normal controls
European Journal of Human Genetics (2020)