Abstract
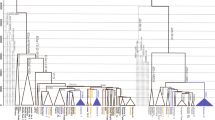
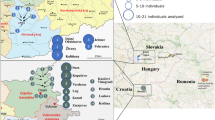
Human Y-chromosome haplogroup structure is largely circumscribed by continental boundaries. One notable exception to this general pattern is the young haplogroup R1a that exhibits post-Glacial coalescent times and relates the paternal ancestry of more than 10% of men in a wide geographic area extending from South Asia to Central East Europe and South Siberia. Its origin and dispersal patterns are poorly understood as no marker has yet been described that would distinguish European R1a chromosomes from Asian. Here we present frequency and haplotype diversity estimates for more than 2000 R1a chromosomes assessed for several newly discovered SNP markers that introduce the onset of informative R1a subdivisions by geography. Marker M434 has a low frequency and a late origin in West Asia bearing witness to recent gene flow over the Arabian Sea. Conversely, marker M458 has a significant frequency in Europe, exceeding 30% in its core area in Eastern Europe and comprising up to 70% of all M17 chromosomes present there. The diversity and frequency profiles of M458 suggest its origin during the early Holocene and a subsequent expansion likely related to a number of prehistoric cultural developments in the region. Its primary frequency and diversity distribution correlates well with some of the major Central and East European river basins where settled farming was established before its spread further eastward. Importantly, the virtual absence of M458 chromosomes outside Europe speaks against substantial patrilineal gene flow from East Europe to Asia, including to India, at least since the mid-Holocene.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Frazer KA, Ballinger DG, Cox DR et al: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Underhill PA, Kivisild T : Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet 2007; 41: 539–564.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Underhill PA, Shen P, Lin AA et al: Y chromosome sequence variation and the history of human populations. Nat Genet 2000; 26: 358–361.
YCC: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Quintana-Murci L, Krausz C, Zerjal T et al: Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am J Hum Genet 2001; 68: 537–542.
Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249.
Gimbutas M : Proto-Indo-European culture. The Kurgan culture during the fifth, fourth, and third millenia B.C.; in Cardona G, Hoenigswald HM, Senn A (eds): Indo-European and Indo-Europeans. Philadelphia: University of Pennsylvania Press, 1970, pp 155–195.
Hinds DA, Stuve LL, Nilsen GB et al: Whole-genome patterns of common DNA variation in three human populations. Science 2005; 307: 1072–1079.
Repping S, van Daalen SK, Brown LG et al: High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet 2006; 38: 463–467.
Gusmão L, Alves C, Costa S et al: Point mutations in the flanking regions of the Y-chromosome specific STRs DYS391, DYS437 and DYS438. Int J Legal Med 2002; 116: 322–326.
Zhivotovsky LA, Underhill PA, Cinnioǧlu C et al: The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 2004; 74: 50–61.
Sengupta S, Zhivotovsky LA, King R et al: Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 2006; 78: 202–221.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ : Iran tricontinental nexus for Y-chromosome driven migration. Hum Hered 2006; 61: 132–143.
Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ : Y-chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet 2008; 16: 374–386.
Stefan M, Stefanescu G, Gavrila L et al: Y chromosome analysis reveals a sharp genetic boundary in the Carpathian region. Eur J Hum Genet 2001; 9: 27–33.
Al-Zahery N, Semino O, Benuzzi G et al: Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 2003; 28: 458–472.
Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J : High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet 2001; 68: 1019–1029.
Deng W, Shi B, He X et al: Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J Hum Genet 2004; 49: 339–348.
Di Giacomo F, Luca F, Anagnou N et al: Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects. Mol Phylogenet Evol 2003; 28: 387–395.
Karafet T, Xu L, Du R et al: Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet 2001; 69: 615–628.
Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF : High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol 2002; 74: 761–789.
Karlsson AO, Wallerström T, Gotherström A, Holmlund G : Y-chromosome diversity in Sweden – a long-time perspective. Eur J Hum Genet 2006; 14: 963–970.
Katoh T, Munkhbat B, Tounai K et al: Genetic features of Mongolian ethnic groups revealed by Y-chromosomal analysis. Gene 2005; 346: 63–70.
Kharkov VN : Structure of Y-chromosomal Lineages in Siberian Populations. Tomsk: Siberian Division of Russian Academy of Medical Sciences, 2005.
Kivisild T, Rootsi S, Metspalu M et al: The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 2003; 72: 313–332.
Lappalainen T, Koivumaki S, Salmela E et al: Regional differences among the Finns: a Y-chromosomal perspective. Gene 2006; 376: 207–215.
Lappalainen T, Laitinen V, Salmela E et al: Migration waves to the Baltic Sea region. Ann Hum Genet 2008; 72: 337–348.
Qamar R, Ayub Q, Mohyuddin A et al: Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 2002; 70: 1107–1124.
Sahoo S, Singh A, Himabindu G et al: A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci USA 2006; 103: 843–848.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 2000; 290: 1155–1159.
Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA : Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet 2002; 70: 265–268.
Zalloua PA, Xue Y, Khalife J et al: Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet 2008; 82: 873–882.
Zerjal T, Wells R, Yuldasheva N, Ruzibakiev R, Tyler-Smith C : A genetic landscape reshaped by recent events: Y-chromosomal insights into Central Asia. Am J Hum Genet 2002; 71: 466–482.
Cinnioǧlu King R, Kivisild T et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Luis JR, Rowold DJ, Regueiro M et al: The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 2004; 74: 532–544.
Mohyuddin A, Ayub Q, Underhill PA, Tyler-Smith C, Mehdi SQ : Detection of novel Y SNPs provides further insights into Y chromosomal variation in Pakistan. J Hum Genet 2006; 51: 375–378.
King RJ, Ozcan SS, Carter T et al: Differential Y-chromosome Anatolian influences on the Greek and Cretan Neolithic. Ann Hum Genet 2008; 72: 205–214.
Kayser M, Lao O, Anslinger K et al: Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum Genet 2005; 117: 428–443.
Woźniak M, Grzybowski T, Starzyñski J, Marciniak T : Continuity of Y chromosome haplotypes in the population of Southern Poland before and after the Second World War. Forensic Sci Int 2007; 1: 134–140.
Weale ME, Yepiskoposyan L, Jager RF et al: Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum Genet 2001; 109: 659–674.
Telegin D, Lillie M, Potekhina I, Kovaliukh M : Settlement and economy in Neolithic Ukraine: a new chronology. Antiquity 2003; 77: 456–470.
Zvelebil M : Innovating Hunter-Gathers. The Mesolithic in the Baltic; in Bailey G, Spikins P (eds): Mesolithic Europe. Cambridge: Cambridge University Press, 2008, pp 18–59.
Price T, Bentley R, Lüning J, Gronenborn D, Wahl J : Prehistoric human migration in the Linearbandkeramik of central Europe. Antiquity 2001; 75: 593–603.
Buchvaldek M : Corded pottery complex in central Europe. J Indo-European Studies 1980; 8: 393–406.
Currat M, Excoffier L : The effect of the Neolithic expansion on European molecular diversity. Proc Biol Sci 2005; 272: 679–688.
Bramanti B, Thomas MG, Haak W et al: Genetic discontinuity between local hunter-gatherers and central Europe's first farmers. Science 2009; 326: 137–140.
Haak W, Forster P, Bramanti B et al: Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science 2005; 310: 1016–1018.
Malyarchuk B, Grzybowski T, Derenko M et al: Mitochondrial DNA phylogeny in Eastern and Western Slavs. Mol Biol Evol 2008; 25: 1651–1658.
Beja-Pereira A, Luikart G, England PR et al: Gene-culture coevolution between cattle milk protein genes and human lactase genes. Nat Genet 2003; 35: 311–313.
Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG : Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci USA 2007; 104: 3736–3741.
Itan Y, Powell A, Beaumont MA, Burger J, Thomas MG : The origins of lactase persistence in Europe. PLoS Comput Biol 2009; 5: e1000491.
Anthony D : The Horse, the Wheel and Language. How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. Princeton, NJ: Princeton University Press, 2007.
Sherratt A : The transformation of early agrarian Europe: the later Neolithic and Copper Ages 4500-2500 BC; in Cunliffe B (ed): Prehistoric Europe: An Illustrated History. Oxford: Oxford University Press, 1998, pp 167–201.
Haak W, Brandt G, de Jong HN et al: Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc Natl Acad Sci USA 2008; 105: 18226–18231.
Gray RD, Atkinson QD : Language-tree divergence times support the Anatolian theory of Indo-European origin. Nature 2003; 426: 435–439.
Acknowledgements
We thank all the men who donated DNA samples used in this study. This research was supported by the European Union European Regional Development Fund through the Centre of Excellence in Genomics, Estonian Biocentre and Tartu University, EC Grant ECOGENE (205419) to Estonian Biocentre, Estonian Science Foundation Grant No. 7445 (to SR) and Estonian Basic Research Grant SF 0270177s08 (to RV), Croatian Ministry of Science, Education and Sports Grant 196-1962766-2751 (to PR), grants of the RAS Programs ‘Origin and Evolution of the Biosphere’ and ‘Molecular and Cell Biology’ to LZ, a grant to OS by V Compagnia di San Paolo and Progetti di Ricerca Interesse Nazionale 2007 (Italian Ministry of the University), and the grant of the Ministry of Education of the Slovak Republic and of Slovak Academy of Sciences – VEGA No. 1/3245/06 to MB. QA, AM and SQM were supported by The Wellcome Trust. We thank Scott R Woodward and the Sorenson Molecular Genealogy Foundation for providing support for AAL and PAU.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Underhill, P., Myres, N., Rootsi, S. et al. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet 18, 479–484 (2010). https://doi.org/10.1038/ejhg.2009.194
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2009.194
Keywords
This article is cited by
-
Contrasting maternal and paternal genetic histories among five ethnic groups from Khyber Pakhtunkhwa, Pakistan
Scientific Reports (2022)
-
The Y chromosome of autochthonous Basque populations and the Bronze Age replacement
Scientific Reports (2021)
-
Subdividing Y-chromosome haplogroup R1a1 reveals Norse Viking dispersal lineages in Britain
European Journal of Human Genetics (2021)
-
Sex-biased patterns shaped the genetic history of Roma
Scientific Reports (2020)
-
Middle eastern genetic legacy in the paternal and maternal gene pools of Chuetas
Scientific Reports (2020)