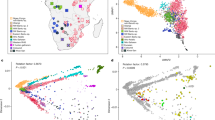
Abstract
Studies of large sets of single nucleotide polymorphism (SNP) data have proven to be a powerful tool in the analysis of the genetic structure of human populations. In this work, we analyze genotyping data for 2841 SNPs in 12 sub-Saharan African populations, including a previously unsampled region of southeastern Africa (Mozambique). We show that robust results in a world-wide perspective can be obtained when analyzing only 1000 SNPs. Our main results both confirm the results of previous studies, and show new and interesting features in sub-Saharan African genetic complexity. There is a strong differentiation of Nilo-Saharans, much beyond what would be expected by geography. Hunter-gatherer populations (Khoisan and Pygmies) show a clear distinctiveness with very intrinsic Pygmy (and not only Khoisan) genetic features. Populations of the West Africa present an unexpected similarity among them, possibly the result of a population expansion. Finally, we find a strong differentiation of the southeastern Bantu population from Mozambique, which suggests an assimilation of a pre-Bantu substrate by Bantu speakers in the region.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Cavalli-Sforza LL, Menozzi P, Piazza A : The History and Geography of Human Genes. Princeton University Press: Princeton, 1994.
Salas A, Richards M, De la Fe T et al: The making of the African mtDNA landscape. Am J Hum Genet 2002; 71: 1082–1111.
Batini C, Coia V, Battaggia C et al: Phylogeography of the human mitochondrial L1c haplogroup: genetic signatures of the prehistory of Central Africa. Mol Phylogenet Evol 2007; 43: 635–644.
Quintana-Murci L, Quach H, Harmant C et al: Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc Natl Acad Sci USA 2008; 105: 1596–1601.
Behar DM, Villems R, Soodyall H et al: The dawn of human matrilineal diversity. Am J Hum Genet 2008; 82: 1130–1140.
Cruciani F, Santolamazza P, Shen P et al: A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 2002; 70: 1197–1214.
Berniell-Lee G, Calafell F, Bosch E et al: Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages. Mol Biol Evol 2009; 26: 1581–1589.
Tishkoff SA, Reed FA, Friedlaender FR et al: The genetic structure and history of Africans and African Americans. Science 2009; 324: 1035–1044.
Bryc K, Auton A, Nelson MR et al: Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA 2010; 107: 786–791.
Li JZ, Absher DM, Tang H et al: Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008; 319: 1100–1104.
Jakobsson M, Scholz SW, Scheet P et al: Genotype, haplotype and copy-number variation in worldwide human populations. Nature 2008; 451: 998–1003.
Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L : Natural selection has driven population differentiation in modern humans. Nat Genet 2008; 40: 340–345.
Patterson N, Price AL, Reich D : Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
Pritchard JK, Stephens M, Donnelly P : Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945–959.
Falush D, Stephens M, Pritchard JK : Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003; 164: 1567–1587.
Novembre J, Stephens M : Interpreting principal component analyses of spatial population genetic variation. Nat Genet 2008; 40: 646–649.
Vansina J : New Linguistic evidence and ‘the bantu expansion’. J African Hist 1995; 36: 173–195.
Phillipson DW : African Archaeology. Cambridge University Press: Cambridge, 1993.
Newman JL : The Peopling of Africa: A Geographic Interpretation. Yale University Press: New Haven, London, 1995.
Oslisly R : The middle Ogooué valley, Gabon: cultural changes and palaeoclimatic implications of the last fourmillenia. Azania 1995; 39–40: 324–331.
Pereira L, Gusmao L, Alves C, Amorim A, Prata MJ : Bantu and European Y-lineages in sub-Saharan Africa. Ann Hum Genet 2002; 66: 369–378.
Pereira L, Macaulay V, Torroni A, Scozzari R, Prata MJ, Amorim A : Prehistoric and historic traces in the mtDNA of Mozambique: insights into the Bantu expansions and the slave trade. Ann Hum Genet 2001; 65: 439–458.
Plaza S, Salas A, Calafell F et al: Insights into the western Bantu dispersal: mtDNA lineage analysis in Angola. Hum Genet 2004; 115: 439–447.
Beleza S, Gusmao L, Amorim A, Carracedo A, Salas A : The genetic legacy of western Bantu migrations. Hum Genet 2005; 117: 366–375.
Acknowledgements
This research was supported by the Dirección General de Investigación, Ministerio de Ciencia y Tecnología, Spain (grants SAF2007-63171, BFU2007-63657) and Direcció General de Recerca, Generalitat de Catalunya (2009 SGR 1101). SNP genotyping services were provided by the Spanish ‘Centro Nacional de Genotipado’ (CEGEN; http://www.cegen.org); Bioinformatic services were kindly provided by the Genomic Diversity node, Spanish Bioinformatic Institute (http://www.inab.org). MS was supported by a PhD fellowship from the Programa de becas FPU del Ministerio de Educación y Ciencia, Spain (AP2005-3982).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Sikora, M., Laayouni, H., Calafell, F. et al. A genomic analysis identifies a novel component in the genetic structure of sub-Saharan African populations. Eur J Hum Genet 19, 84–88 (2011). https://doi.org/10.1038/ejhg.2010.141
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2010.141
Keywords
This article is cited by
-
The influence of habitats on female mobility in Central and Western Africa inferred from human mitochondrial variation
BMC Evolutionary Biology (2013)
-
Prediction of people’s origin from degraded DNA—presentation of SNP assays and calculation of probability
International Journal of Legal Medicine (2013)