Abstract
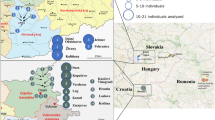
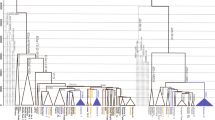
The phylogenetic relationships of numerous branches within the core Y-chromosome haplogroup R-M207 support a West Asian origin of haplogroup R1b, its initial differentiation there followed by a rapid spread of one of its sub-clades carrying the M269 mutation to Europe. Here, we present phylogeographically resolved data for 2043 M269-derived Y-chromosomes from 118 West Asian and European populations assessed for the M412 SNP that largely separates the majority of Central and West European R1b lineages from those observed in Eastern Europe, the Circum-Uralic region, the Near East, the Caucasus and Pakistan. Within the M412 dichotomy, the major S116 sub-clade shows a frequency peak in the upper Danube basin and Paris area with declining frequency toward Italy, Iberia, Southern France and British Isles. Although this frequency pattern closely approximates the spread of the Linearbandkeramik (LBK), Neolithic culture, an advent leading to a number of pre-historic cultural developments during the past ≤10 thousand years, more complex pre-Neolithic scenarios remain possible for the L23(xM412) components in Southeast Europe and elsewhere.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Barbujani G, Bertorelle G : Genetics and the population history of Europe. Proc Natl Acad Sci USA 2001; 98: 22–25.
Underhill PA, Kivisild T : Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet 2007; 41: 539–564.
Green RE, Krause J, Briggs AW et al: A draft sequence of the Neandertal genome. Science 2010; 328: 710–722.
Novelletto A : Y chromosome variation in Europe: continental and local processes in the formation of the extant gene pool. Ann Hum Biol 2007; 34: 139–172.
Jobling MA, Tyler-Smith C : The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 2003; 4: 598–612.
Chiaroni J, Underhill PA, Cavalli-Sforza LL : Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci USA 2009; 106: 20174–20179.
Cinnioğlu C, King R, Kivisild T et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ : Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered 2006; 61: 132–143.
Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249.
Sengupta S, Zhivotovsky LA, King R et al: Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 2006; 78: 202–221.
Sahoo S, Singh A, Himabindu G et al: A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci USA 2006; 103: 843–848.
Bereir RE, Hassan HY, Salih NA et al: Co-introgression of Y-chromosome haplogroups and the sickle cell gene across Africa's Sahel. Eur J Hum Genet 2007; 15: 1183–1185.
Cruciani F, Trombetta B, Sellitto D et al: Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur J Hum Genet 2010; 18: 800–807.
Kayser M, Lao O, Anslinger K et al: Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum Genet 2005; 117: 428–443.
Underhill PA : Inferring Human History: Clues from Y-Chromosome Haplotypes. Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press: NY, USA, 2003, Vol LXVIII, pp 487–493.
Alonso S, Flores C, Cabrera V et al: The place of the Basques in the European Y-chromosome diversity landscape. Eur J Hum Genet 2005; 13: 1293–1302.
Battaglia V, Fornarino S, Al-Zahery N et al: Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur J Hum Genet 2009; 17: 820–830.
Al-Zahery N, Semino O, Benuzzi G et al: Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 2003; 28: 458–472.
Robino C, Crobu F, Di Gaetano C et al: Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample. Int J Legal Med 2008; 122: 251–255.
Rosser ZH, Zerjal T, Hurles ME et al: Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 2000; 67: 1526–1543.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 2000; 290: 1155–1159.
Arredi B, Poloni ES, Tyler-Smith C : The Peopling of Europe. Anthropological Genetics: Theory, Methods and Applications. Cambridge University Press: Cambridge, 2010.
Balaresque P, Bowden GR, Adams SM et al: A predominantly neolithic origin for European paternal lineages. PLoS Biol 2010; 8: e1000285.
Sims LM, Garvey D, Ballantyne J : Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs. Hum Mutat 2007; 28: 97.
Myres NM, Ekins JE, Lin AA, Cavalli-Sforza LL, Woodward SR, Underhill PA : Y-chromosome short tandem repeat DYS458.2 non-consensus alleles occur independently in both binary haplogroups J1-M267 and R1b3-M405. Croat Med J 2007; 48: 450–459.
Hinds DA, Stuve LL, Nilsen GB et al: Whole-genome patterns of common DNA variation in three human populations. Science 2005; 307: 1072–1079.
Repping S, van Daalen SK, Brown LG et al: High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet 2006; 38: 463–467.
Underhill PA, Myres NM, Rootsi S et al: Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet 2009; 18: 479–484.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Zhivotovsky LA, Underhill PA, Cinnioğlu C et al: The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 2004; 74: 50–61.
Pichler I, Fuchsberger C, Platzer C et al: Drawing the history of the Hutterite population on a genetic landscape: inference from Y-chromosome and mtDNA genotypes. Eur J Hum Genet 2010; 18: 463–470.
Trivedi R, Sahoo S, Singh A et al: Genetic imprints of Pleistocene origin of Indian populations: a comprehensive Phylogeographic sketch of Indian Y-chromosomes. Int J Hum Genet 2008; 8: 97–118.
Shi W, Ayub Q, Vermeulen M et al: A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Mol Biol Evol 2010; 27: 385–393.
Whittle A : The first farmers; In: Cunliffe B (ed): Prehistoric Europe: An Illustrated History. Oxford University Press: Oxford, 1998, pp 136–166.
Burkill M : The Middle Neolithic of the Paris Basin and North-Eastern France; In: Scarre C (ed).: Ancient France, Neolithic Societies and Their Landscapes 6000-2000 BC. The Edinburgh University Press: Edinburgh, 1983, p 394.
Sherratt A : The transformation of early agrarian Europe: the later Neolithic and Copper Ages 4500-2500 BC; In: Cunliffe B (ed): Prehistoric Europe: An Illustrated History. Oxford University Press: Oxford, 1998, pp 167–201.
Collard M, Edinborough K, Shennan S, Thomas M : Radiocarbon evidence indicates that migrants introduced farming to Britain. J Arch Sci 2009; 37: 866–870.
Perlès C : The Early Neolithic in Greece: The First Farming Communities in Europe. Cambridge: University of Cambridge Press, 2001, p 395.
Price TD : Europe's First Farmers: An Introduction: Europe's First Farmers. Cambridge University Press: Cambridge, 2000, pp 1–18.
Edmonds CA, Lillie AS, Cavalli-Sforza LL : Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci USA 2004; 101: 975–979.
Klopfstein S, Currat M, Excoffier L : The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol 2006; 23: 482–490.
Zimmermann A, Hilpert J, Wendt KP : Estimations of population density for selected periods between the Neolithic and AD 1800. Hum Biol 2009; 81: 357–380.
Cuniliffe B : Iron Age societies in Western Europe and beyond, 900-140 BC; In: Cunliffe B (ed): Prehistoric Europe: An Illustrated History. Oxford University Press: Oxford, 1998, pp 336–372.
Adams SM, King TE, Bosch E, Jobling MA : The case of the unreliable SNP: recurrent back-mutation of Y-chromosomal marker P25 through gene conversion. Forensic Sci Int 2006; 159: 14–20.
Acknowledgements
We thank all the men who donated DNA samples used in this study. We thank Vincent Vizachero for alerting us to the potential of M520, M529, S116, L11, L23 SNPs and for insight regarding the DYS390, 19 repeat allele and M73. Professor Joachim Hallmayer provided laboratory facilities for PAU and AAL. This research was supported by the European Union European Regional Development Fund through the Centre of Excellence in Genomics, Estonian Biocentre and Tartu University, Estonian Science Foundation Grant no. 7445 (to SR) and Estonian Basic Research Grant SF 0270177s08 (to RV) and by the Artur Lind scholarship from the Estonian Genome Foundation (to MJ) and the Croatian Ministry of Science, Education and Sports Grant 196-1962766-2751 (to PR) and the grants of the Russian foundation for basic research 10-06-00451 and 10-07-00515 to OB and AP. We thank Scott R Woodward and the Sorenson Molecular Genealogy Foundation for providing support for AAL and PAU. RJK was supported by a grant from the Stanford-France Interdisciplinary Program. JC was supported by a grant from the ANR Project AFGHAPOP, no. BLAN07-3_222301, CSD 9.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Myres, N., Rootsi, S., Lin, A. et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet 19, 95–101 (2011). https://doi.org/10.1038/ejhg.2010.146
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2010.146
Keywords
This article is cited by
-
Large-scale pedigree analysis highlights rapidly mutating Y-chromosomal short tandem repeats for differentiating patrilineal relatives and predicting their degrees of consanguinity
Human Genetics (2023)
-
Population genetic study of 17 Y-STR Loci of the Sorani Kurds in the Province of Sulaymaniyah, Iraq
BMC Genomics (2022)
-
Y-chromosome target enrichment reveals rapid expansion of haplogroup R1b-DF27 in Iberia during the Bronze Age transition
Scientific Reports (2022)
-
Development and validation of a new multiplex for upgrading Y-STRs population databases from 12 to 23 markers and its forensic casework application
Scientific Reports (2022)
-
Contrasting maternal and paternal genetic histories among five ethnic groups from Khyber Pakhtunkhwa, Pakistan
Scientific Reports (2022)