Abstract
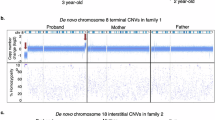
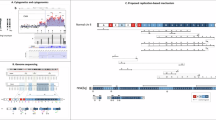
We evaluated 966 consecutive pediatric patients with various developmental disorders by high-resolution microarray-based comparative genomic hybridization and found 10 individuals with pathogenic copy number variants (CNVs) on the short arm of chromosome 8 (8p), representing approximately 1% of the patients analyzed. Two patients with 8p terminal deletion associated with interstitial inverted duplication (inv dup del(8p)) had different mechanisms leading to the formation of a dicentric intermediate during meiosis. Three probands carried an identical ∼5.0 Mb interstitial duplication of chromosome 8p23.1. Four possible hotspots within 8p were observed at nucleotide coordinates of ∼10.45, 24.32–24.82, 32.19–32.77, and 38.94–39.72 Mb involving the formation of recurrent genomic rearrangements. Other CNVs with deletion- or duplication-specific start or stop coordinates on the 8p provide useful information for exploring the basic mechanisms of complex structural rearrangements in the human genome.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Nusbaum C, Mikkelsen TS, Zody MC et al: DNA sequence and analysis of human chromosome 8. Nature 2006; 439: 331–335.
Vermeesch JR, Thoelen R, Salden I, Raes M, Matthijs G, Fryns JP : Mosaicism del(8p)/inv dup(8p) in a dysmorphic female infant: a mosaic formed by a meiotic error at the 8p OR gene and an independent terminal deletion event. J Med Genet 2003; 40: e93.
Giorda R, Ciccone R, Gimelli G et al: Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat 2007; 28: 459–468.
Hollox EJ, Barber JC, Brookes AJ, Armour JA : Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res 2008; 18: 1686–1697.
Cooke SL, Northup JK, Champaige NL et al: Molecular cytogenetic characterization of a unique and complex de novo 8p rearrangement. Am J Med Genet A 2008; 146A: 1166–1172.
Shimokawa O, Miyake N, Yoshimura T et al: Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am J Med Genet A 2005; 136: 49–51.
Barber JC, Maloney VK, Huang S et al: 8p23.1 duplication syndrome; a novel genomic condition with unexpected complexity revealed by array CGH. Eur J Hum Genet 2008; 16: 18–27.
Giglio S, Broman KW, Matsumoto N et al: Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 2001; 68: 874–883.
Devriendt K, Matthijs G, Van Dael R et al: Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am J Hum Genet 1999; 64: 1119–1126.
Shimokawa O, Kurosawa K, Ida T et al: Molecular characterization of inv dup del(8p): analysis of five cases. Am J Med Genet A 2004; 128A: 133–137.
Graw SL, Sample T, Bleskan J, Sujansky E, Patterson D : Cloning, sequencing, and analysis of inv8 chromosome breakpoints associated with recombinant 8 syndrome. Am J Hum Genet 2000; 66: 1138–1144.
Weleber RG, Verma RS, Kimberling WJ, Fieger Jr HG, lubs HA : Duplication-deficiency of the short arm of chromosome 8 following artificial insemination. Ann Genet 1976; 19: 241–247.
Buysse K, Antonacci F, Callewaert B et al: Unusual 8p inverted duplication deletion with telomere capture from 8q. Eur J Med Genet 2009; 52: 31–36.
Floridia G, Piantanida M, Minelli A et al: The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am J Hum Genet 1996; 58: 785–796.
Pabst B, Arslan-Kirchner M, Schmidtke J, Miller K : The application of region-specific probes for the resolution of duplication 8p: a case report and a review of the literature. Cytogenet Genome Res 2003; 103: 3–7.
Dill FJ, Schertzer M, Sandercock J, Tischler B, Wood S : Inverted tandem duplication generates a duplication deficiency of chromosome 8p. Clin Genet 1987; 32: 109–113.
Barber JC, James RS, Patch C, Temple IK : Protelomeric sequences are deleted in cases of short arm inverted duplication of chromosome 8. Am J Med Genet 1994; 50: 296–299.
Minelli A, Floridia G, Rossi E et al: D8S7 is consistently deleted in inverted duplications of the short arm of chromosome 8 (inv dup 8p). Hum Genet 1993; 92: 391–396.
Guo WJ, Callif-Daley F, Zapata MC, Miller ME : Clinical and cytogenetic findings in seven cases of inverted duplication of 8p with evidence of a telomeric deletion using fluorescence in situ hybridization. Am J Med Genet 1995; 58: 230–236.
de Die-Smulders CE, Engelen JJ, Schrander-Stumpel CT et al: Inversion duplication of the short arm of chromosome 8: clinical data on seven patients and review of the literature. Am J Med Genet 1995; 59: 369–374.
Macmillin MD, Suri V, Lytle C, Krauss CM : Prenatal diagnosis of inverted duplicated 8p. Am J Med Genet 2000; 93: 94–98.
Felbor U, Knotgen N, Schams G, Buwe A, Steinlein C, Schmid M : Mosaicism for an ectopic NOR at 8pter and a complex rearrangement of chromosome 8 in a patient with severe psychomotor retardation. Cytogenet Genome Res 2004; 106: 55–60.
Zuffardi O, Bonaglia M, Ciccone R, Giorda R : Inverted duplications deletions: underdiagnosed rearrangements? Clin Genet 2009; 75: 505–513.
Rowe LR, Lee JY, Rector L et al: U-type exchange is the most frequent mechanism for inverted duplication with terminal deletion rearrangements. J Med Genet 2009; 46: 694–702.
Zollino M, Murdolo M, Marangi G et al: On the nosology and pathogenesis of Wolf–Hirschhorn syndrome: genotype–phenotype correlation analysis of 80 patients and literature review. Am J Med Genet C Semin Med Genet 2008; 148C: 257–269.
Ozkinay F, Kanit H, Onay H et al: Prenatal diagnosis of de novo unbalanced translocation 8p;21q using subtelomeric probes. Genet Couns 2006; 17: 315–320.
Liehr T, Mrasek K, Weise A et al: Small supernumerary marker chromosomes – progress towards a genotype–phenotype correlation. Cytogenet Genome Res 2006; 112: 23–34.
Fellermann K, Stange DE, Schaeffeler E et al: A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet 2006; 79: 439–448.
Cho SC, Yim SH, Yoo HK et al: Copy number variations associated with idiopathic autism identified by whole-genome microarray-based comparative genomic hybridization. Psychiatr Genet 2009; 19: 177–185.
Hollox EJ, Huffmeier U, Zeeuwen PL et al: Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet 2008; 40: 23–25.
Yu S, Bittel DC, Kibiryeva N, Zwick DL, Cooley LD : Validation of the Agilent 244K oligonucleotide array-based comparative genomic hybridization platform for clinical cytogenetic diagnosis. Am J Clin Pathol 2009; 132: 349–360.
Yu S, Kielt M, Stegner AL, Kibiryeva N, Bittel DC, Cooley LD : Quantitative real-time polymerase chain reaction for the verification of genomic imbalances detected by microarray-based comparative genomic hybridization. Genet Test Mol Biomarkers 2009; 13: 751–760.
Shinawi M, Cheung SW : The array CGH and its clinical applications. Drug Discov Today 2008; 13: 760–770.
Ciccone R, Mattina T, Giorda R et al: Inversion polymorphisms and non-contiguous terminal deletions: the cause and the (unpredicted) effect of our genome architecture. J Med Genet 2006; 43: e19.
Ballif BC, Gajecka M, Shaffer LG : Monosomy 1p36 breakpoints indicate repetitive DNA sequence elements may be involved in generating and/or stabilizing some terminal deletions. Chromosome Res 2004; 12: 133–141.
Ballif BC, Wakui K, Gajecka M, Shaffer LG : Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Hum Genet 2004; 114: 198–206.
Chabchoub E, Rodriguez L, Galan E et al: Molecular characterisation of a mosaicism with a complex chromosome rearrangement: evidence for coincident chromosome healing by telomere capture and neo-telomere formation. J Med Genet 2007; 44: 250–256.
Knijnenburg J, van Haeringen A, Hansson KB et al: Ring chromosome formation as a novel escape mechanism in patients with inverted duplication and terminal deletion. Eur J Hum Genet 2007; 15: 548–555.
Kostiner DR, Nguyen H, Cox VA, Cotter PD : Stabilization of a terminal inversion duplication of 8p by telomere capture from 18q. Cytogenet Genome Res 2002; 98: 9–12.
Kennedy SJ, Teebi AS, Adatia I, Teshima I : Inherited duplication, dup (8) (p23.1p23.1) pat, in a father and daughter with congenital heart defects. Am J Med Genet 2001; 104: 79–80.
Tsai CH, Graw SL, McGavran L : 8p23 duplication reconsidered: is it a true euchromatic variant with no clinical manifestation? J Med Genet 2002; 39: 769–774.
Barber JC, Maloney V, Hollox EJ et al: Duplications and copy number variants of 8p23.1 are cytogenetically indistinguishable but distinct at the molecular level. Eur J Hum Genet 2005; 13: 1131–1136.
Slavotinek A, Lee SS, Davis R et al: Fryns syndrome phenotype caused by chromosome microdeletions at 15q26.2 and 8p23.1. J Med Genet 2005; 42: 730–736.
Devriendt K, De Mars K, De Cock P, Gewillig M, Fryns JP : Terminal deletion in chromosome region 8p23.1-8pter in a child with features of velo-cardio-facial syndrome. Ann Genet 1995; 38: 228–230.
Barber JC, Joyce CA, Collinson MN et al: Duplication of 8p23.1: a cytogenetic anomaly with no established clinical significance. J Med Genet 1998; 35: 491–496.
Engelen JJ, Moog U, Evers JL, Dassen H, Albrechts JC, Hamers AJ : Duplication of chromosome region 8p23.1 → p23.3: a benign variant? Am J Med Genet 2000; 91: 18–21.
Glancy M, Barnicoat A, Vijeratnam R et al: Transmitted duplication of 8p23.1–8p23.2 associated with speech delay, autism and learning difficulties. Eur J Hum Genet 2009; 17: 37–43.
Higgins AW, Alkuraya FS, Bosco AF et al: Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet 2008; 82: 712–722.
Willemsen MH, de Leeuw N, Pfundt R, de Vries BB, Kleefstra T : Clinical and molecular characterization of two patients with a 6.75 Mb overlapping deletion in 8p12p21 with two candidate loci for congenital heart defects. Eur J Med Genet 2009; 52: 134–139.
Klopocki E, Fiebig B, Robinson P et al: A novel 8 Mb interstitial deletion of chromosome 8p12–p21.2. Am J Med Genet A 2006; 140: 873–877.
Bettio D, Baldwin EL, Carrozzo R et al: Molecular cytogenetic and clinical findings in a patient with a small supernumerary r(8) mosaicism. Am J Med Genet A 2008; 146A: 247–250.
Makoff AJ, Flomen RH : Detailed analysis of 15q11–q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader–Willi, Angelman, and inv dup(15) syndromes. Genome Biol 2007; 8: R114.
Lupski JR : Structural variation in the human genome. N Engl J Med 2007; 356: 1169–1171.
Gu W, Zhang F, Lupski JR : Mechanisms for human genomic rearrangements. Pathogenetics 2008; 1: 4.
de Smith AJ, Walters RG, Coin LJ et al: Small deletion variants have stable breakpoints commonly associated with alu elements. PLoS One 2008; 3: e3104.
Hastings PJ, Ira G, Lupski JR : A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 2009; 5: e1000327.
Acknowledgements
We thank Dr Holly H Ardinger for the critical review and comments on the article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, S., Fiedler, S., Stegner, A. et al. Genomic profile of copy number variants on the short arm of human chromosome 8. Eur J Hum Genet 18, 1114–1120 (2010). https://doi.org/10.1038/ejhg.2010.66
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2010.66
Keywords
This article is cited by
-
Mechanisms of structural chromosomal rearrangement formation
Molecular Cytogenetics (2022)
-
Prenatal and postnatal diagnoses and phenotype of 8p23.3p22 duplication in one family
BMC Medical Genomics (2021)
-
The involvement of U-type dicentric chromosomes in the formation of terminal deletions with or without adjacent inverted duplications
Human Genetics (2020)
-
Identification of candidate genes for congenital heart defects on proximal chromosome 8p
Scientific Reports (2016)
-
Genotype–Phenotype Association Studies of Chromosome 8p Inverted Duplication Deletion Syndrome
Behavior Genetics (2011)