Abstract
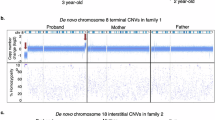
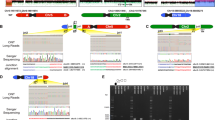
High-resolution genome-wide array analysis enables detailed screening for cryptic and submicroscopic imbalances of microscopically balanced de novo rearrangements in patients with developmental delay and/or congenital abnormalities. In this report, we added the results of genome-wide array analysis in 54 patients to data on 117 patients from seven other studies. A chromosome imbalance was detected in 37% of all patients with two-breakpoint rearrangements. In 49% of these patients, the imbalances were located in one or both breakpoint regions. Imbalances were more frequently (90%) found in complex rearrangements, with the majority (81%) having deletions in the breakpoint regions. The size of our own cohort enabled us to relate the presence of an imbalance to the clinical features of the patients by using a scoring system, the De Vries criteria, that indicates the complexity of the phenotype. The median De Vries score was significantly higher (P=0.002) in those patients with an imbalance (5, range 1–9) than in patients with a normal array result (3, range 0–7). This study provides accurate percentages of cryptic imbalances that can be detected by genome-wide array analysis in simple and complex de novo microscopically balanced chromosome rearrangements and confirms that these imbalances are more likely to occur in patients with a complex phenotype.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Jacobs PA, Browne C, Gregson N, Joyce C, White H : Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet 1992; 29: 103–108.
Gardner RJMS, Sutherland GR : Chromosome Abnormalities and Genetic Counseling, 3rd edn. Oxford University Press: New York, 2004.
Warburton D : De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet 1991; 49: 995–1013.
Rauch A, Hoyer J, Guth S et al: Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A 2006; 140: 2063–2074.
Hochstenbach R, van Binsbergen E, Engelen J et al: Array analysis and karyotyping: workflow consequences based on a retrospective study of 36 325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet 2009; 52: 161–169.
Miller DT, Adam MP, Aradhya S et al: Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010; 86: 749–764.
Stankiewicz P, Beaudet AL : Use of genome-wide array analysis in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev 2007; 17: 182–192.
Vissers LE, de Vries BB, Veltman JA : Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet 2010; 47: 289–297.
Gribble SM, Prigmore E, Burford DC et al: The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J Med Genet 2005; 42: 8–16.
De Gregori M, Ciccone R, Magini P et al: Cryptic deletions are a common finding in ‘balanced’ reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet 2007; 44: 750–762.
Sismani C, Kitsiou-Tzeli S, Ioannides M et al: Cryptic genomic imbalances in patients with de novo or familial apparently balanced translocations and abnormal phenotype. Mol Cytogenet 2008; 1: 15.
Baptista J, Mercer C, Prigmore E et al: Breakpoint mapping and genome-wide array analysis in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet 2008; 82: 927–936.
Higgins AW, Alkuraya FS, Bosco AF et al: Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet 2008; 82: 712–722.
Schluth-Bolard C, Delobel B, Sanlaville D et al: Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet 2009; 52: 291–296.
Gijsbers ACJ, Bosch CAJ, Dauwerse JG et al: Additional cryptic CNVs in mentally retarded patients with apparently balanced karyotypes. Eur J Med Genet 2010; 53: 227–233.
Baptista J, Prigmore E, Gribble SM, Jacobs PA, Carter NP, Crolla JA : Molecular cytogenetic analyses of breakpoints in apparently balanced reciprocal translocations carried by phenotypically normal individuals. Eur J Hum Genet 2005; 13: 1205–1212.
de Vries BB, White SM, Knight SJ et al: Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet 2001; 38: 145–150.
de Vries BB, Pfundt R, Leisink M et al: Diagnostic genome profiling in mental retardation. Am J Hum Genet 2005; 77: 606–616.
McMullan DJ, Bonin M, Hehir-Kwa JY et al: Molecular karyotyping of patients with unexplained mental retardation by SNP arrays: a multicenter study. Hum Mutat 2009; 30: 1082–1092.
Nannya Y, Sanada M, Nakazaki K et al: A robust algorithm for copy number detection using high-density oligonucleotide single-nucleotide polymorphism genotyping arrays. Cancer Res 2005; 65: 6071–6079.
Hehir-Kwa J, Egmont-Petersen M, Janssen I, Smeets D, van Kessel A, Veltman J : Genome-wide copy number profiling on high-density bacterial artificial chromosomes, single-nucleotide polymorphisms, and oligonucleotide microarrays: a platform comparison based on statistical power analysis. DNA Res 2007; 14: 1–11.
Hilhortst-Hofstee Y, Hamel BCJ, Verheij JBGM et al: The clinical spectrum of complete FBN1 allele deletions. Eur J Hum Genet 2011; 19: 247–252.
Shaikh TH, Gai X, Perin JC et al: High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res 2009; 19: 1682–1690.
Howarth KD, Pole JCM, Beavis JC et al: Large duplications at reciprocal translocation breakpoints that might be the counterpart of large deletions and could arise from stalled replication bubbles. Genome Res 2011; 21: 525–534.
Mefford HC, Muhle H, Ostertag P et al: Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet 2010; 6: e1000962.
de Kovel CG, Trucks H, Helbig I et al: Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain 2010; 133: 23–32.
Mefford HC, Sharp AJ, Baker C et al: Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 2008; 359: 1685–1699.
Hanemaaijer N, Dijkhuizen T, Haadsma M et al: A 649 kb microduplication in 1p34.1, including POMGNT1, in a patient with microcephaly, coloboma and laryngomalacia; and a review of the literature. Eur J Med Genet 2009; 52: 116–119.
Kalscheuer VM, Feenstra I, van Ravenswaaij-Arts CM et al: Disruption of the TCF4 gene in a girl with mental retardation but without the classical Pitt-Hopkins syndrome. Am J Med Genet A 2008; 146: 2053–2059.
Chen W, Ullmann R, Langnick C et al: Breakpoint analysis of balanced chromosome rearrangements by next-generation paired-end sequencing. Eur J Hum Genet 2010; 18: 539–543.
van Silfhout AT, van den Akker PC, Dijkhuizen T et al: Split hand/foot malformation due to chromosome 7q aberrations (SHFM1): additional support for functional haploinsufficiency as the causative mechanism. Eur J Hum Genet 2009; 17: 1432–1438.
Verhoeven WMA, Tuerlings JHAM, van Ravenswaay-Arts CMA, Boermans JAJ, Tuinier S : Chromosome abnormalities in clinical psychiatry: a report of two older patients. Eur J Psychiatry 2007; 21: 207–211.
Verhoeven WMA, van Ravenswaaij-Arts CMA, de Leeuw N et al: Disturbed serine metabolism and psychosis in a patient with a de novo translocation (2;10)(p23;q22.1). Genet Couns 2006; 17: 421–428.
Concannon N, Hegarty AM, Stallings RL, Reardon W : Coffin-Lowry phenotype in a patient with a complex chromosome rearrangement. J Med Genet 2002; 39: e41.
Acknowledgements
We are grateful to all the patients and their parents for their kind cooperation. We would also like to thank the requesting physicians, Bregje van Bon, Ineke van de Burgt, Ton van Essen, Ben Hamel, Marjolijn Jongmans, Tjitske Kleefstra, Carlo Marcelis, Sonja de Munnik, Gretel Oudesluijs, C Nur Semerci, Liesbeth Spruijt, Irene Stolte-Dijkstra, Peter van Tintelen, Joep Tuerlings and Michèl Willemsen, for their contribution. Special thanks to the Array Diagnostics teams of Groningen and Nijmegen, to Hanneke Mieloo for extensive FISH analyses, to Marian Bakker for statistical assistance and to Jackie Senior for editorial support. This work was supported by grants from the Netherlands Organization for Health Research and Development (ZonMW 917-86-319 to BdV) and the Brain Foundation of the Netherlands (Hersenstichting) (BdV)
Web resources
The URLs for data presented here are as follows:
Database of Genomic Variants (DGV), http://projects.tcag.ca/variation/
DECIPHER database, http://decipher.sanger.ac.uk/
European Cytogeneticists Association Register for Unbalanced Chromosome Aberrations (ECARUCA), http://ecaruca.net
Ensembl Human Genome Browser, http://www.ensembl.org/Homo_sapiens/
Online Mendelian Inheritance in Man, http://www.ncbi.nlm.nih.gov/Omim
University of California-Santa Cruz Human Genome Browser, http://genome.ucsc.edu/
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Feenstra, I., Hanemaaijer, N., Sikkema-Raddatz, B. et al. Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur J Hum Genet 19, 1152–1160 (2011). https://doi.org/10.1038/ejhg.2011.120
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2011.120
Keywords
This article is cited by
-
Evaluation of genetic risk of apparently balanced chromosomal rearrangement carriers by breakpoint characterization
Journal of Assisted Reproduction and Genetics (2024)
-
Identification of a familial complex chromosomal rearrangement by optical genome mapping
Molecular Cytogenetics (2022)
-
The phenotypic spectrum and genotype-phenotype correlations in 106 patients with variants in major autism gene CHD8
Translational Psychiatry (2022)
-
Mosaicism for structural non-centromeric autosomal rearrangements in disease-defined carriers: sex differences in the rearrangements profile and maternal age distributions
Molecular Cytogenetics (2017)
-
Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases
Nature Genetics (2017)