Abstract
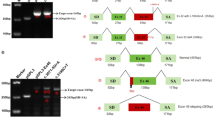
UK NHS diagnostic service sequence analysis of genes generally examines and reports on variations within a designated region 5′ and 3′ of each exon, typically 30 bp up and downstream. However, because of the degenerate nature of the splice sites, intronic variants outside the AG and GT dinucleotides of the acceptor and donor splice sites (ASS and DSS) are most often classified as being of unknown clinical significance, unless there is some functional evidence of their pathogenicity. It is now becoming clear that mutations deep within introns can also interfere with normal processing of pre-mRNA and result in pathogenic effects on the mature transcript. In diagnostic laboratories, these deep intronic variants most often fall outside of the regions analysed and so are rarely reported. With the likelihood that next generation sequencing will identify more of these unclassified variants, it will become important to perform additional studies to determine the pathogenicity of such sequence anomalies. Here, we analyse variants detected in either COL2A1 or COL11A1 in patients with Stickler syndrome. These have been analysed both in silico and functionally using either RNA isolated from the patient's cells or, more commonly, minigenes as splicing reporters. We show that deep intronic mutations are not a rare occurrence, including one variant that results in multiple transcripts, where both de novo donor and ASS are created by the mutation. Another variant produces transcripts that result in either haploinsufficiency or a dominant negative effect, potentially modifying the disease phenotype.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Patthy L : Modular assembly of genes and the evolution of new functions. Genetica 2003; 118: 217–231.
Liu M, Grigoriev A : Protein domains correlate strongly with exons in multiple eukaryotic genomes-evidence of exon shuffling? Trends in Genet 2004; 20: 399–403.
Modrek B, Lee C : A genomic view of alternative splicing. Nat Genet 2002; 30: 13–19.
Boue S, Letunic I, Bork P : Alternative splicing and evolution. Bioessays 2003; 25: 1031–1034.
Wang G-S, Cooper TA : Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 2007; 8: 749–761.
Baralle D, Lucassen A, Buratti E : Missed threads. EMBO R 2009; 10: 810–816.
Ahmad NN, Ala-Kokko L, Knowlton RG et al: Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-ophthalmopathy). Proc Natl Acad Sci USA 1991; 88: 6624–6627.
Richards AJ, Yates JRW, Williams R et al: A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in α1(XI) collagen. Hum Mol Genet 1996; 5: 1339–1343.
Annunen S, Korkko J, Czarny M et al: Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome, but other mutations cause overlapping Marshall/Stickler phenotypes. Am J Hum Genet 1999; 65: 974–983.
Richards AJ, Laidlaw M, Whittaker J et al: High efficiency of mutation detection in type 1 stickler syndrome using a two-stage approach: vitreoretinal assessment coupled with exon sequencing for screening COL2A1. Hum Mut 2006; 27: 696–704.
Richards AJ, McNinch A, Martin H et al: Stickler syndrome and the vitreous phenotype: mutations in COL2A1 and COL11A1. Hum Mut 2010; 31: E1461–E1471.
Hoornaert KP, Vereecke I, Dewinter C et al: Stickler syndrome caused by COL2A1 mutations: genotype-phenotype correlation in a series of 100 patients. Eur J Hum Genet 2010; 18: 872–880.
Snead MP, Yates JRW : Clinical and Molecular Genetics of Stickler syndrome. J Med Genet 1999; 36: 353–359.
Carroll C, Papaioannou D, Rees A et al: The clinical effectiveness and safety of prophylactic retinal interventions to reduce the risk of retinal detachment and subsequent vision loss in adults and children with Stickler syndrome: a systematic review. Health Technol Assess 2011; 15: 1–62.
Yaguchi H, Ikeda T, Osada H et al: Identification of the COL2A1 mutation in patients with type I Stickler syndrome using RNA from freshly isolated peripheral white blood cells. Genet Test Mol Biomarkers 2011; 15: 231–237.
Spranger J, Winterpacht A, Zabel B : Kniest dysplasia: Dr. W. Kniest, his patient, the molecular defect. Am J Med Genet 1997; 69: 79–84.
Richards AJ, Snead MP : The influence of pre-mRNA splicing on phenotypic modification in Stickler's syndrome and other type II collagenopathies. Eye 2008; 22: 1243–1250.
Martin S, Richards AJ, Yates JRW et al: Stickler syndrome: further mutations in COL11A1 and evidence for additional locus heterogeneity. Eur J Hum Genet 1999; 7: 807–814.
Gaildrat P, Killian A, Martins A et al: Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol Biol 2010; 653: 249–257.
Limb GA, Salt TE, Munro PM et al: In vitro characterization of a spontaneously immortalized human Müller cell line (MIO-M1). Invest Ophthalmol Vis Sci 2002; 43: 864–869.
Richards AJ, Meredith S, Poulson A et al: A novel mutation of COL2A1 resulting in dominantly inherited rhegmatogenous retinal detachment. Invest Ophthalmol Vis Sci 2005; 46: 663–668.
Ang A, Poulson AV, Goodburn SF et al: Retinal detachment and prophylaxis in type 1 Stickler syndrome. Ophthalmology 2008; 115: 164–168.
Winterpacht A, Hilbert M, Schwarze U et al: Kniest and Stickler dysplasia phenotypes caused by collagen type II gene (COL2A1) defect. Nat Genet 1993; 3: 323–326.
Voelkerding KV, Dames SA, Durtschi JD : Next-generation sequencing: from basic research to diagnostics. Clin Chem 2009; 55: 641–658.
Kuhlenbaumer G, Hullmann J, Appenzeller S : Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mut 2011; 32: 144–151.
Acknowledgements
We thank all the staff in the Molecular Genetics Department at Addenbrooke's Hospital for their contribution to this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors are commissioned by the Advisory Group for National Specialised Services (http://www.specialisedservices.nhs.uk) and the Department of Health to provide a national diagnostic service for Stickler syndrome.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Richards, A., McNinch, A., Whittaker, J. et al. Splicing analysis of unclassified variants in COL2A1 and COL11A1 identifies deep intronic pathogenic mutations. Eur J Hum Genet 20, 552–558 (2012). https://doi.org/10.1038/ejhg.2011.223
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2011.223
Keywords
This article is cited by
-
Impact of rs11024102 PLEKHA7, rs3753841 COL11A1 single nucleotide polymorphisms, and serum levels of oxidative stress markers on the risk of primary angle-closure glaucoma in Egyptians
Journal of Genetic Engineering and Biotechnology (2022)
-
Deep intronic mutations and human disease
Human Genetics (2017)
-
A novel COL11A1 mutation affecting splicing in a patient with Stickler syndrome
Human Genome Variation (2015)
-
Dawn of ocular gene therapy: implications for molecular diagnosis in retinal disease
Science China Life Sciences (2013)