Abstract
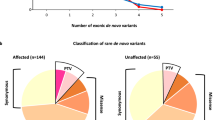
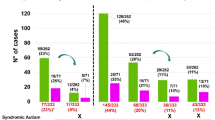
Individuals with autism are more likely to carry rare inherited and de novo copy number variants (CNVs). However, further research is needed to establish which CNVs are causal and the mechanisms by which these CNVs influence autism. We examined genomic DNA of children with autism (N=41) and healthy controls (N=367) for rare CNVs using a high-resolution array comparative genomic hybridization platform. We show that individuals with autism are more likely to harbor rare CNVs as small as ∼10 kb, a threshold not previously detectable, and that CNVs in cases disproportionately affect genes involved in transcription, nervous system development, and receptor activity. We also show that a subset of genes that have known or suspected allele-specific or imprinting effects and are within rare-case CNVs may undergo loss of transcript expression. In particular, expression of CNTNAP2 and ZNF214 are decreased in probands compared with their unaffected transmitting parents. Furthermore, expression of PRODH and ARID1B, two genes affected by de novo CNVs, are decreased in probands compared with controls. These results suggest that for some genes affected by CNVs in autism, reduced transcript expression may be a mechanism of pathogenesis during neurodevelopment.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bailey A, Le Couteur A, Gottesman I et al: Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 1995; 25: 63–77.
Autism Genome Project Consortium: Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 2007; 39: 319–328.
Christian SL, Brune CW, Sudi J et al: Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry 2008; 63: 1111–1117.
Glessner JT, Wang K, Cai G et al: Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009; 459: 569–573.
Guilmatre A, Dubourg C, Mosca A-L et al: Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry 2009; 66: 947–956.
Marshall C, Noor A, Vincent J et al: Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 2008; 82: 477–488.
Morrow EM, Yoo S-Y, Flavell SW et al: Identifying autism loci and genes by tracing recent shared ancestry. Science 2008; 321: 218–223.
Pinto D, Pagnamenta AT, Klei L et al: Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466: 368–372.
Sebat J, Lakshmi B, Malhotra D et al: Strong association of de novo copy number mutations with autism. Science 2007; 316: 445–449.
Weiss LA, Shen Y, Korn JM et al: Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 2008; 358: 667–675.
Abrahams BS, Geschwind DH : Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 2008; 9: 341–355.
Hogart A, Wu D, Lasalle JM, Schanen NC : The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis 2008; 46: 86–93.
Cook Jr EH, Scherer SW : Copy-number variations associated with neuropsychiatric conditions. Nature 2008; 455: 919–923.
King BH, Hollander E, Sikich L et al: Lack of efficacy of citalopram in children with autism spectrum disorders and high levels of repetitive behavior: citalopram ineffective in children with autism. Arch Gen Psychiatry 2009; 66: 583–590.
Talati A, Fyer AJ, Weissman MM : A comparison between screened NIMH and clinically interviewed control samples on neuroticism and extraversion. Mol Psychiatry 2008; 13: 122–130.
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Mi H, Guo N, Kejariwal A, Thomas PD : PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res 2007; 35: D247–D252.
Girirajan S, Rosenfeld JA, Cooper GM et al: A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 2010; 42: 203–209.
Nagl Jr NG, Wang X, Patsialou A, Van Scoy M, Moran E : Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J 2007; 26: 752–763.
Koga M, Ishiguro H, Yazaki S et al: Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet 2009; 18: 2483–2494.
Meechan DW, Tucker ES, Maynard TM, LaMantia A-S : Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc Natl Acad Sci USA 2009; 106: 16434–16445.
Alders M, Ryan A, Hodges M et al: Disruption of a novel imprinted zinc-finger gene, ZNF215, in Beckwith-Wiedemann syndrome. Am J Hum Genet 2000; 66: 1473–1484.
Kent L, Bowdin S, Kirby GA, Cooper WN, Maher ER : Beckwith Weidemann syndrome: a behavioral phenotype-genotype study. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 1295–1297.
Alarcón M, Abrahams BS, Stone JL et al: Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 2008; 82: 150–159.
Arking DE, Cutler DJ, Brune CW et al: A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet 2008; 82: 160–164.
Poot M, Beyer V, Schwaab I et al: Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics 2010; 11: 81–89.
Herzing LBK, Cook EH, Ledbetter DH : Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11- q13 duplications. Hum Mol Genet 2002; 11: 1707–1718.
Bah J, Quach H, Ebstein RP et al: Maternal transmission disequilibrium of the glutamate receptor GRIK2 in schizophrenia. Neuroreport 2004; 15: 1987–1991.
Acknowledgements
This work was supported by the National Institutes of Health grants R01MH083989, R01MH076431, R01HG004222, U54MH066398, U01HD045023, U54MH066673, U54MH068172, U54MH066418, U54MH066494, 5T32ES007032, and T32ES015459.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Supplementary information
Appendix
Appendix
Bryan H King, MD, Seattle Children's Hospital, University of Washington, Seattle, WA; Eric Hollander, MD, Institute of Clinical Neuroscience, New York, NY; Linmarie Sikich, MD University of North Carolina, Chapel Hill, NC; James T McCracken, MD, University of California at Los Angeles, Los Angeles, CA; Lawrence Scahill MSN, PhD, Yale University, New Haven, CT; Joel D Bregman, MD, North Shore–Long Island Jewish Health System, Great Neck, NY; Craig L Donnelly, MD, Dartmouth Medical School, Hanover, NH; Kimberly Dukes, PhD, Boston University, Boston, MA; Deborah Hirtz, MD, National Institute of Neurological Disorders and Stroke, Bethesda, MD; Ann Wagner, PhD, National Institute of Mental Health, Rockville, MD.
Rights and permissions
About this article
Cite this article
Nord, A., Roeb, W., Dickel, D. et al. Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur J Hum Genet 19, 727–731 (2011). https://doi.org/10.1038/ejhg.2011.24
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2011.24
Keywords
This article is cited by
-
Neurobiology of ARID1B haploinsufficiency related to neurodevelopmental and psychiatric disorders
Molecular Psychiatry (2022)
-
Early postnatal serotonin modulation prevents adult-stage deficits in Arid1b-deficient mice through synaptic transcriptional reprogramming
Nature Communications (2022)
-
Role of JAK-STAT and PPAR-Gamma Signalling Modulators in the Prevention of Autism and Neurological Dysfunctions
Molecular Neurobiology (2022)
-
Neuroanatomy and behavior in mice with a haploinsufficiency of AT-rich interactive domain 1B (ARID1B) throughout development
Molecular Autism (2021)
-
Identification of primary copy number variations reveal enrichment of Calcium, and MAPK pathways sensitizing secondary sites for autism
Egyptian Journal of Medical Human Genetics (2020)