Abstract
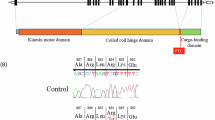
The hereditary spastic paraplegias (HSPs) are a clinically and genetically heterogeneous group of neurodegenerative diseases characterised by progressive spasticity in the lower limbs. The nosology of autosomal recessive forms is complex as most mapped loci have been identified in only one or a few families and account for only a small percentage of patients. We used next-generation sequencing focused on the SPG30 chromosomal region on chromosome 2q37.3 in two patients from the original linked family. In addition, wide genome scan and candidate gene analysis were performed in a second family of Palestinian origin. We identified a single homozygous mutation, p.R350G, that was found to cosegregate with the disease in the SPG30 kindred and was absent in 970 control chromosomes while affecting a strongly conserved amino acid at the end of the motor domain of KIF1A. Homozygosity and linkage mapping followed by mutation screening of KIF1A allowed us to identify a second mutation, p.A255V, in the second family. Comparison of the clinical features with the nature of the mutations of all reported KIF1A families, including those reported recently with hereditary sensory and autonomic neuropathy, suggests phenotype–genotype correlations that may help to understand the mechanisms involved in motor neuron degeneration. We have shown that mutations in the KIF1A gene are responsible for SPG30 in two autosomal recessive HSP families. In published families, the nature of the KIF1A mutations seems to be of good predictor of the underlying phenotype and vice versa.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Depienne C, Stevanin G, Brice A, Durr A : Hereditary spastic paraplegias: an update. Curr Opin Neurol 2007; 20: 674–680.
Salinas S, Proukakis C, Crosby A, Warner TT : Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol 2008; 7: 1127–1138.
Stevanin G, Ruberg M, Brice A : Recent advances in the genetics of spastic paraplegias. Curr Neurol Neurosci Rep 2008; 8: 198–210.
Erlich Y, Edvardson S, Hodges E et al: Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res 2011; 21: 658–664.
Riviere JB, Ramalingam S, Lavastre V et al: KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet 2011.
Klebe S, Azzedine H, Durr A et al: Autosomal recessive spastic paraplegia (SPG30) with mild ataxia and sensory neuropathy maps to chromosome 2q37.3. Brain 2006; 129: 1456–1462.
Al-Bassam J, Cui Y, Klopfenstein D, Carragher BO, Vale RD, Milligan RA : Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J Cell Biol 2003; 163: 743–753.
Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD : The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 1997; 90: 959–966.
Rice S, Lin AW, Safer D et al: A structural change in the kinesin motor protein that drives motility. Nature 1999; 402: 778–784.
Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA : KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett 2011; 491: 168–173.
Zahn TR, Angleson JK, MacMorris MA et al: Dense core vesicle dynamics in Caenorhabditis elegans neurons and the role of kinesin UNC-104. Traffic 2004; 5: 544–559.
Yonekawa Y, Harada A, Okada Y et al: Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol 1998; 141: 431–441.
Goizet C, Boukhris A, Mundwiller E et al: Complicated forms of autosomal dominant hereditary spastic paraplegia are frequent in SPG10. Hum Mutat 2009; 30: E376–E385.
Reid E, Kloos M, Ashley-Koch A et al: A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am J Hum Genet 2002; 71: 1189–1194.
Dion PA, Daoud H, Rouleau GA : Genetics of motor neuron disorders: new insights into pathogenic mechanisms. Nat Rev Genet 2009; 10: 769–782.
Arnoldi A, Crimella C, Tenderini E et al: Clinical phenotype variability in patients with hereditary spastic paraplegia type 5 associated with CYP7B1 mutations. Clin Genet 2011.
De Michele G, De Fusco M, Cavalcanti F et al: A new locus for autosomal recessive hereditary spastic paraplegia maps to chromosome 16q24.3. Am J Hum Genet 1998; 63: 135–139.
Goizet C, Boukhris A, Durr A et al: CYP7B1 mutations in pure and complex forms of hereditary spastic paraplegia type 5. Brain 2009; 132: 1589–1600.
Heinzlef O, Paternotte C, Mahieux F et al: Mapping of a complicated familial spastic paraplegia to locus SPG4 on chromosome 2p. J Med Genet 1998; 35: 89–93.
Ribai P, Stevanin G, Bouslam N et al: A new phenotype linked to SPG27 and refinement of the critical region on chromosome. J Neurol 2006; 253: 714–719.
Hewamadduma C, McDermott C, Kirby J et al: New pedigrees and novel mutation expand the phenotype of REEP1-associated hereditary spastic paraplegia (HSP). Neurogenetics 2009; 10: 105–110.
Elleuch N, Depienne C, Benomar A et al: Mutation analysis of the paraplegin gene (SPG7) in patients with hereditary spastic paraplegia. Neurology 2006; 66: 654–659.
Tsaousidou MK, Ouahchi K, Warner TT et al: Sequence alterations within CYP7B1 implicate defective cholesterol homeostasis in motor-neuron degeneration. Am J Hum Genet 2008; 82: 510–515.
Stevanin G, Azzedine H, Denora P et al: Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain 2008; 131: 772–784.
Schüle R, Kremer BP, Kassubek J et al: SPG10 is a rare cause of spastic paraplegia in European families. J Neurol Neurosurg Psychiatry 2008; 79: 584–587.
Acknowledgements
We are grateful to the DNA and cell bank of the CR-ICM for technical assistance, Drs Bouslam and Nelson for their help and Drs P Bastien, J Melki, V Meiner and M Koenig for patient referral and clinical evaluation. This study was supported financially by grants from the Agence Nationale de la Recherche (ANR SPAX to AD; E-Rare EUROSPA to AB), the Verum Foundation (to AB), the Chief Scientist, MOH, Israel/E-Rare 2007 (to AL) and the GIS-Rare Diseases Institute (NGS call: ARHSP to AB). SK received fellowships from the post-doctoral programme of the German Academic Exchange Service (DAAD, Germany), the Verum Foundation and the TWS foundation (Muenster, Germany)’.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Klebe, S., Lossos, A., Azzedine, H. et al. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur J Hum Genet 20, 645–649 (2012). https://doi.org/10.1038/ejhg.2011.261
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2011.261
Keywords
This article is cited by
-
Illustration of a rare case of hereditary spastic paraplegia type 30 associated with a missense variant in the non-motor domain of KIF1A
Journal of Neurology (2022)
-
Monoallelic KIF1A-related disorders: a multicenter cross sectional study and systematic literature review
Journal of Neurology (2022)
-
Challenges and Controversies in the Genetic Diagnosis of Hereditary Spastic Paraplegia
Current Neurology and Neuroscience Reports (2021)
-
KIF1A-related autosomal dominant spastic paraplegias (SPG30) in Russian families
BMC Neurology (2020)
-
KIF1A variants are a frequent cause of autosomal dominant hereditary spastic paraplegia
European Journal of Human Genetics (2020)