Abstract
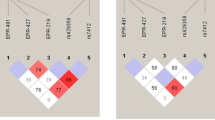
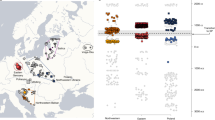
Population isolates have long been of interest to genetic epidemiologists because of their potential to increase power to detect disease-causing genetic variants. The Sorbs of Germany are considered as cultural and linguistic isolates and have recently been the focus of disease association mapping efforts. They are thought to have settled in their present location in eastern Germany after a westward migration from a largely Slavic-speaking territory during the Middle Ages. To examine Sorbian genetic diversity within the context of other European populations, we analyzed genotype data for over 30 000 autosomal single-nucleotide polymorphisms from over 200 Sorbs individuals. We compare the Sorbs with other European individuals, including samples from population isolates. Despite their geographical proximity to German speakers, the Sorbs showed greatest genetic similarity to Polish and Czech individuals, consistent with the linguistic proximity of Sorbian to other West Slavic languages. The Sorbs also showed evidence of subtle levels of genetic isolation in comparison with samples from non-isolated European populations. The level of genetic isolation was less than that observed for the Sardinians and French Basque, who were clear outliers on multiple measures of isolation. The finding of the Sorbs as only a minor genetic isolate demonstrates the need to genetically characterize putative population isolates, as they possess a wide range of levels of isolation because of their different demographic histories.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Heutink P, Oostra BA : Gene finding in genetically isolated populations. Hum Mol Genet 2002; 11: 2507–2515.
Kristiansson K, Naukkarinen J, Peltonen L : Isolated populations and complex disease gene identification. Genome Biol 2008; 9: 109.
Kruglyak L : Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 1999; 22: 139–144.
Eaves IA, Merriman TR, Barber RA et al: The genetically isolated populations of Finland and sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet 2000; 25: 320–323.
Dunning AM, Durocher F, Healey CS et al: The extent of linkage disequilibrium in four populations with distinct demographic histories. Am J Hum Genet 2000; 67: 1544–1554.
Service S, DeYoung J, Karayiorgou M et al: Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet 2006; 38: 556–560.
Angius A, Hyland FC, Persico I et al: Patterns of linkage disequilibrium between SNPs in a Sardinian population isolate and the selection of markers for association studies. Hum Hered 2008; 65: 9–22.
Ethnologue: Languages of the World; Vol. Sixteenth. SIL International Online, http://www.ethnologue.com/.
Behar DM, Thomas MG, Skorecki K et al: Multiple origins of Ashkenazi levites: y chromosome evidence for both near Eastern and European ancestries. Am J Hum Genet 2003; 73: 768–779.
Rodig H, Grum M, Grimmecke HD : Population study and evaluation of 20 Y-chromosome STR loci in Germans. Int J Legal Med 2007; 121: 24–27.
Immel UD, Krawczak M, Udolph J et al: Y-chromosomal STR haplotype analysis reveals surname-associated strata in the East-German population. Eur J Hum Genet 2006; 14: 577–582.
Krawczak M, Lu TT, Willuweit S, Roewer L : Genetic diversity in the German population. in: Cooper D, Kehrer-Sawatzki H (eds): Handbook of Human Molecular Evolution. Wiley: Chichester, 2008, Vol 12, pp 451–456.
Bottcher Y, Unbehauen H, Kloting N et al: Adipose tissue expression and genetic variants of the bone morphogenetic protein receptor 1A gene (BMPR1A) are associated with human obesity. Diabetes 2009; 58: 2119–2128.
Schleinitz D, Carmienke S, Bottcher Y et al: Role of genetic variation in the cannabinoid type 1 receptor gene (CNR1) in the pathophysiology of human obesity. Pharmacogenomics 2010; 11: 693–702.
Tonjes A, Koriath M, Schleinitz D et al: Genetic variation in GPR133 is associated with height: genome wide association study in the self-contained population of Sorbs. Hum Mol Genet 2009; 18: 4662–4668.
Tonjes A, Zeggini E, Kovacs P et al: Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur J Hum Genet 2010; 18: 104–110.
Hoffmann K, Planitz C, Ruschendorf F et al: A novel locus for arterial hypertension on chromosome 1p36 maps to a metabolic syndrome trait cluster in the Sorbs, a Slavic population isolate in Germany. J Hypertens 2009; 27: 983–990.
Nelson MR, Bryc K, King KS et al: The population reference sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet 2008; 83: 347–358.
Novembre J, Johnson T, Bryc K et al: Genes mirror geography within Europe. Nature 2008.
Cann HM, de Toma C, Cazes L et al: A human genome diversity cell line panel. Science 2002; 296: 261–262.
Li JZ, Absher DM, Tang H et al: Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008; 319: 1100–1104.
Arcos-Burgos M, Muenke M : Genetics of population isolates. Clin Genet 2002; 61: 233–247.
Cavalli-Sforza LL, Menozzi P, Piazza A : The History and Geography of Human Genes. New Jersey: Princeton University Press, 1994.
Roberts DF : Who are the Orcadians? Anthropol Anz 1986; 44: 93–104.
Roberts DF : Genetic affinities of the Shetland islanders. Ann Hum Biol 1990; 17: 121–132.
Patterson N, Price AL, Reich D : Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
McVean G : A genealogical interpretation of principal components analysis. PLoS Genet 2009; 5: e1000686.
Weir BS, Cockerham CC : Estimating F-statistics for the analysis of population structure. Evolution 1984; 38: 1358–1370.
Alexander DH, Novembre J, Lange K : Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19: 1655–1664.
Jakobsson M, Scholz SW, Scheet P et al: Genotype, haplotype and copy-number variation in worldwide human populations. Nature 2008; 451: 998–1003.
Pickrell JK, Coop G, Novembre J et al: Signals of recent positive selection in a worldwide sample of human populations. Genome Res 2009; 19: 826–837.
Passarino G, Underhill PA, Cavalli-Sforza LL et al: Y chromosome binary markers to study the high prevalence of males in Sardinian centenarians and the genetic structure of the Sardinian population. Hum Hered 2001; 52: 136–139.
Calafell F, Bertranpetit J : Principal component analysis of gene frequencies and the origin of Basques. Am J Phys Anthropol 1994; 93: 201–215.
Geary PJ : The myth of nations: the medieval origins of Europe. Princeton: Princeton University Press, 2002.
Lao O, Lu TT, Nothnagel M et al: Correlation between genetic and geographic structure in Europe. Curr Biol 2008; 18: 1241–1248.
Tenesa A, Wright AF, Knott SA et al: Extent of linkage disequilibrium in a Sardinian sub-isolate: sampling and methodological considerations. Hum Mol Genet 2004; 13: 25–33.
Bosch E, Laayouni H, Morcillo-Suarez C et al: Decay of linkage disequilibrium within genes across HGDP-CEPH human samples: most population isolates do not show increased LD. BMC Genomics 2009; 10: 338.
Garagnani P, Laayouni H, Gonzalez-Neira A et al: Isolated populations as treasure troves in genetic epidemiology: the case of the Basques. Eur J Hum Genet 2009; 17: 1490–1494.
Laayouni H, Calafell F, Bertranpetit J : A genome-wide survey does not show the genetic distinctiveness of Basques. Hum Genet 2010; 127: 455–458.
Rodriguez-Ezpeleta N, varez-Busto J, Imaz L et al: High-density SNP genotyping detects homogeneity of Spanish and French Basques, and confirms their genomic distinctiveness from other European populations. Hum Genet 2010; 128: 113–117.
Price AL, Helgason A, Palsson S et al: The impact of divergence time on the nature of population structure: an example from Iceland. PLoS Genet 2009; 5: e1000505.
O’Dushlaine C, McQuillan R, Weale ME et al: Genes predict village of origin in rural Europe. Eur J Hum Genet 2010; 18: 1269–1270.
Acknowledgements
The authors thank all sample donors, Knut Krohn (Microarray Core Facility of the Interdisciplinary Centre for Clinical Research, University of Leipzig) for the genotyping support, and Mark I McCarthy and Nigel William Rayner for analytical and bioinformatics support. Financial support was received from the Andrew W Mellon Foundation (KV), Searle Scholars Program (JN), German Research Council (KFO-152), IZKF (B27) and the German Diabetes Association. AG and MSc were funded by the Leipzig Interdisciplinary Research Cluster of Genetic Factors, Clinical Phenotypes and Environment (LIFE Center, University of Leipzig). LIFE is funded by means of the European Union, by the European Regional Development Fund (ERFD) and by means of the Free State of Saxony within the framework of the excellence initiative. IK and DG were funded by grants from the Slovak Diabetes Association and the ERDF (Transendogen/26240220051).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Veeramah, K., Tönjes, A., Kovacs, P. et al. Genetic variation in the Sorbs of eastern Germany in the context of broader European genetic diversity. Eur J Hum Genet 19, 995–1001 (2011). https://doi.org/10.1038/ejhg.2011.65
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2011.65
Keywords
This article is cited by
-
Genome-wide meta-analysis of phytosterols reveals five novel loci and a detrimental effect on coronary atherosclerosis
Nature Communications (2022)
-
Genetically programmed changes in transcription of the novel progranulin regulator
Journal of Molecular Medicine (2020)
-
Metabolic effects of genetic variation in the human REPIN1 gene
International Journal of Obesity (2019)
-
On the impact of relatedness on SNP association analysis
BMC Genetics (2017)
-
Overcoming the dichotomy between open and isolated populations using genomic data from a large European dataset
Scientific Reports (2017)