Abstract
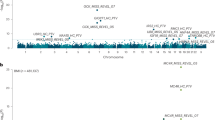
We hypothesize that imputation based on data from the 1000 Genomes Project can identify novel association signals on a genome-wide scale due to the dense marker map and the large number of haplotypes. To test the hypothesis, the Wellcome Trust Case Control Consortium (WTCCC) Phase I genotype data were imputed using 1000 genomes as reference (20100804 EUR), and seven case/control association studies were performed using imputed dosages. We observed two ‘missed’ disease-associated variants that were undetectable by the original WTCCC analysis, but were reported by later studies after the 2007 WTCCC publication. One is within the IL2RA gene for association with type 1 diabetes and the other in proximity with the CDKN2B gene for association with type 2 diabetes. We also identified two refined associations. One is SNP rs11209026 in exon 9 of IL23R for association with Crohn's disease, which is predicted to be probably damaging by PolyPhen2. The other refined variant is in the CUX2 gene region for association with type 1 diabetes, where the newly identified top SNP rs1265564 has an association P-value of 1.68 × 10−16. The new lead SNP for the two refined loci provides a more plausible explanation for the disease association. We demonstrated that 1000 Genomes-based imputation could indeed identify both novel (in our case, ‘missed’ because they were detected and replicated by studies after 2007) and refined signals. We anticipate the findings derived from this study to provide timely information when individual groups and consortia are beginning to engage in 1000 genomes-based imputation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
WTCCC: Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature 2007; 447: 661–678.
Orho-Melander M, Melander O, Guiducci C et al: Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes 2008; 57: 3112–3121.
de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF : Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet 2008; 17: R122–R128.
Li Y, Willer C, Sanna S, Abecasis G : Genotype imputation. Annu Rev Genomics Hum Genet 2009; 10: 387–406.
Marchini J, Howie B : Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511.
Thorisson GA, Smith AV, Krishnan L, Stein LD : The International HapMap Project Web site. Genome Res 2005; 15: 1592–1593.
Durbin RM, Abecasis GR, Altshuler DL et al: A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073.
Liu JZ, Tozzi F, Waterworth DM et al: Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 2010; 42: 436–440.
Sanna S, Pitzalis M, Zoledziewska M et al: Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet 2010; 42: 495–497.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR : MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834.
Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA : Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 2004; 74: 106–120.
Browning BL, Browning SR : A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 2009; 84: 210–223.
Devlin B, Roeder K : Genomic control for association studies. Biometrics 1999; 55: 997–1004.
The International HapMap Consortium: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Pe'er I, Yelensky R, Altshuler D, Daly MJ : Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 2008; 32: 381–385.
Barrett JC, Clayton DG, Concannon P et al: Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009; 41: 703–707.
Saxena R, Voight BF, Lyssenko V et al: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007; 316: 1331–1336.
Shea J, Agarwala V, Philippakis AA et al: Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nat Genet 2011; 43: 801–805.
Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA : Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development 2008; 135: 729–741.
Barrett JC, Hansoul S, Nicolae DL et al: Genome-wide association defines more than 30 distinct susceptibility loci for Crohn′s disease. Nat Genet 2008; 40: 955–962.
Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A : A comprehensive evaluation of SNP genotype imputation. Hum Genet 2009; 125: 163–171.
Pei YF, Li J, Zhang L, Papasian CJ, Deng HW : Analyses and comparison of accuracy of different genotype imputation methods. PLoS One 2008; 3: e3551.
Zheng J, Li Y, Abecasis GR, Scheet P : A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet Epidemiol 2011; 35: 102–110.
Pei YF, Zhang L, Li J, Deng HW : Analyses and comparison of imputation-based association methods. PLoS One 2010; 5: e10827.
Nyholt DR : A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 2004; 74: 765–769.
Conneely KN, Boehnke M : So many correlated tests, so little time! Rapid adjustment of p values for multiple correlated tests. Am J Hum Genet 2007; 81: 1158–1168.
Gao XY : Multiple testing corrections for imputed SNPs. Genet. Epidemiol 2011; 35: 154–158.
Wen SH, Lu ZS : Factors affecting the effective number of tests in genetic association studies: a comparative study of three PCA-based methods. J Hum Genet 2011; 56: 428–435.
Kullo IJ, de Andrade M, Boerwinkle E, McConnell JP, Kardia SL, Turner ST : Pleiotropic genetic effects contribute to the correlation between HDL cholesterol, triglycerides, and LDL particle size in hypertensive sibships. Am J Hypertens 2005; 18: 99–103.
Avery CL, He Q, North KE et al: A phenomics-based strategy identifies loci on APOC1, BRAP, and PLCG1 associated with metabolic syndrome phenotype domains. PLoS Genet 2011; 7: e1002322.
Zawistowski M, Gopalakrishnan S, Ding J, Li Y, Grimm S, Zollner S : Extending rare-variant testing strategies: analysis of noncoding sequence and imputed genotypes. Am J Hum Genet 2010; 87: 604–617.
Acknowledgements
We thank Prof David P Strachan at the St George's University of London for commenting on an earlier version of this manuscript. We acknowledge the WTCC for making the data available. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The effort of DE and AF was supported by the Deutsche Forschungsgemeinschaft (DFG), grant no. FR 2821/2-1, and the German Ministry of Education and Research (BMBF) through the National Genome Research Network (NGFN). This project received infrastructure support through the DFG cluster of excellence ‘Inflammation at Interfaces’. YL is partially supported by the NIH grant R01-HG006292 and 3-R01-CA082659-11S1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Huang, J., Ellinghaus, D., Franke, A. et al. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur J Hum Genet 20, 801–805 (2012). https://doi.org/10.1038/ejhg.2012.3
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2012.3
Keywords
This article is cited by
-
A genome-wide cross-trait analysis identifies shared loci and causal relationships of type 2 diabetes and glycaemic traits with polycystic ovary syndrome
Diabetologia (2022)
-
How imputation can mitigate SNP ascertainment Bias
BMC Genomics (2021)
-
Prediction of functional microexons by transfer learning
BMC Genomics (2021)
-
Impact of pre- and post-variant filtration strategies on imputation
Scientific Reports (2021)
-
Revealing potential drug-disease-gene association patterns for precision medicine
Scientometrics (2021)