Abstract
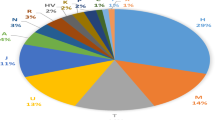
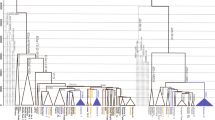
Central Asia has served as a corridor for human migrations providing trading routes since ancient times. It has functioned as a conduit connecting Europe and the Middle East with South Asia and far Eastern civilizations. Therefore, the study of populations in this region is essential for a comprehensive understanding of early human dispersal on the Eurasian continent. Although Y- chromosome distributions in Central Asia have been widely surveyed, present-day Afghanistan remains poorly characterized genetically. The present study addresses this lacuna by analyzing 190 Pathan males from Afghanistan using high-resolution Y-chromosome binary markers. In addition, haplotype diversity for its most common lineages (haplogroups R1a1a*-M198 and L3-M357) was estimated using a set of 15 Y-specific STR loci. The observed haplogroup distribution suggests some degree of genetic isolation of the northern population, likely due to the Hindu Kush mountain range separating it from the southern Afghans who have had greater contact with neighboring Pathans from Pakistan and migrations from the Indian subcontinent. Our study demonstrates genetic similarities between Pathans from Afghanistan and Pakistan, both of which are characterized by the predominance of haplogroup R1a1a*-M198 (>50%) and the sharing of the same modal haplotype. Furthermore, the high frequencies of R1a1a-M198 and the presence of G2c-M377 chromosomes in Pathans might represent phylogenetic signals from Khazars, a common link between Pathans and Ashkenazi groups, whereas the absence of E1b1b1a2-V13 lineage does not support their professed Greek ancestry.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Dupree L, Lawrence JA, Brill RH et al: Prehistoric research in Afghanistan (1959–1966). Trans Am Philos Soc 1972; 62: 1–84.
Katzir I : The Israeli source of the Pathan tribes. The Scribe 2001; 74: 53–54.
Runion ML : The history of Afghanistan. Connecticut Greenwood Press, 2007.
Caroe OP : The Pathans 550 BC-AD 1957. Oxford Oxford University Press, 1976.
Mohyuddin A, Ayub Q, Qamar R et al: Y-chromosomal STR haplotypes in Pakistani populations. Forensic Sci Int 2001; 118: 141–146.
Qamar R, Ayub Q, Mohyuddin A et al: Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 2002; 70: 1107–1124.
Mansoor A, Mazhar K, Khaliq S et al: Investigation of the Greek ancestry of populations from northern Pakistan. Hum Genet 2004; 114: 484–490.
Sengupta S, Zhivotovsky LA, King R et al: Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveals minor genetic influence of Central Asian pastoralists. Am J Hum Genet 2006; 78: 202–221.
Firasat S, Khaliq S, Mohyuddin A et al: Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan. Eur J Hum Genet 2007; 15: 121–126.
Di Cristofaro J, Buhler S, Temori SA, Chiaroni J : Genetic data of 15 STR loci in five populations from Afghanistan. Forensic Sci Int Genet 2012; 6: e44–e45.
Noor S, Ali S, Eaaswarkhanth M, Haque I : Allele frequency distribution for 15 autosomal STR loci in Afridi Pathan population of Uttar Pradesh, India. Leg Med 2009; 11: 308–311.
Antunez-de-Mayolo G, Antunez-de-Mayolo A, Antunez-de-Mayolo P et al: Phylogenetics of worldwide human populations as determined by polymorphic Alu insertions. Electrophoresis 2002; 23: 3346–3356.
Sambrook J, Russell DW : Molecular cloning: A Laboratory Manual. New York Cold Spring Harbor Laboratory Press, 2001.
Gayden T, Regueiro M, Martinez L, Cadenas AM, Herrera RJ : Human Y-chromosome haplotyping by allele-specific polymerase chain reaction. Electrophoresis 2008; 29: 2419–2423.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Underhill PA, Myres NM, Rootsi S et al: Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet 2010; 18: 479–484.
Myres NM, Rootsi S, Lin AA et al: A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet 2011; 19: 95–101.
Lacau H, Bukhari A, Gayden T et al: Y-STR profiling in two Afghanistan populations. Leg Med 2011; 13: 103–108.
Battaglia V, Fornarino S, Al-Zahery N et al: Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur J Hum Genet 2009; 17: 820–830.
Martinez L, Underhill PA, Zhivotovsky LA et al: Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau. Eur J Hum Genet 2007; 15: 485–493.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ : Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered 2006; 61: 132–143.
Behar DM, Garrigan D, Kaplan ME et al: Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum Genet 2004; 114: 354–365.
Rohlf F : NTSYSpc. Setauket, NY Exter Publishing, 2002.
Felsenstein J : PHYLIP-phylogeny inference package (version 3.2). Cladistics 1989; 5: 164–166.
Excoffier L, Laval G, Schneider S : Arlequin ver 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 2005; 1: 47–50.
Slatkin M : A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995; 139: 457–462.
Goodman SJ : Rst Calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and determining their significance. Mol Ecol 1997; 6: 881–885.
Bandelt HJ, Forster P, Röhl A : Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48.
Zhivotovsky LA, Underhill PA, Cinnioğlu C et al: The effective mutation rate at Y chromosome short tandem repeats, with application to human population divergence time. Am J Hum Genet 2004; 74: 50–61.
Goedbloed M, Vermeulen M, Fang RN et al: Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFlSTR Yfiler PCR amplification kit. Int J Legal Med 2009; 123: 471–482.
Kayser M, Krawczak M, Excoffier L et al: An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet 2001; 68: 990–1018.
Klyosov A : DNA genealogy, mutation rates, and some historical evidence written in the Y-chromosome part II: walking the map. J Genet Genealogy 2009; 5: 217–256.
Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian heartland: a continental perspective on Y-chromosomal diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y-chromosome perspective. Science 2000; 290: 1155–1159.
Mirabal S, Regueiro M, Cadenas AM et al: Y-chromosome distribution within the geo-linguistic landscape of northwestern Russia. Eur J Hum Genet 2009; 17: 1260–1273.
Klyosov A : DNA genealogy, mutation rates, and some historical evidence written in the Y-chromosome part I: basic principles and the method. J Genet Genealogy 2009; 5: 186–216.
Wells RS : Deep ancestry: inside the genography project. The landmark DNA quest to decipher our distant past. National Geographics 2007, p. 103.
Shen P, Lavi T, Kivisild T et al: Reconstruction of patrilineages and matrilineages of Samaritans and other Israeli populations from Y-Chromosome and mitochondrial DNA sequence Variation. Hum Mutat 2004; 24: 248–260.
King RJ, DiCristofaro J, Kouvatsi A et al: The coming of the Greeks to provence and corsica: Y-chromosome models of archaic Greek colonization of the western Mediterranean. BMC Evol Biol 2011; 11: 69.
Behar DM, Thomas MG, Skorecki K et al: Multiple origins of Ashkenazi Levites: Y chromosome evidence for both Near Eastern and European ancestries. Am J Hum Genet 2003; 73: 768–779.
Nebel A, Filon D, Faerman M, Soodyall H, Oppenheim A : Y chromosome evidence for a founder effect in Ashkenazi Jews. Eur J Hum Genet 2005; 13: 388–391.
Hammer MF, Behar DM, Karafet TM et al: Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood. Hum Genet 2009; 126: 707–717.
Gayden T, Cadenas AM, Regueiro M et al: The Himalayas as a directional barrier to gene flow. Am J Hum Genet 2007; 80: 884–894.
Nasidze I, Sarkisian T, Kerimov A, Toneking M : Testing hypotheses of language replacement in the Caucasus: evidence from the Y chromosome. Hum Genet 2003; 112: 255–261.
Cinnioğlu C, King R, Kivisild T et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Derenko M, Malyarhuk B, Denisova GA et al: Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum Genet 2006; 118: 591–604.
Fechner A, Quinque D, Rychkov S et al: Boundaries and clines in the West Eurasian Y-chromosome landscape: insights from the European part of Russia. Am J Phys Anthropol 2008; 137: 41–47.
Abu-Amero KK, Hellani A, González AM, Larruga JM, Cabrera VM, Underhill PA : Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions. BMC Genet 2009; 10: 59.
Zalloua PA, Xue Y, Khalife J et al: Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet 2008; 82: 873–882.
Luis JR, Rowold DJ, Regueiro M et al: The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 2004; 74: 532–544.
Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ : Y-chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet 2008; 16: 374–386.
Acknowledgements
The authors express their appreciation to the original DNA donors who made this study possible. We gratefully acknowledge Viviana Arce, Priscilla Torres, David Perez and Silvia Calderon for their contribution to the manuscript and we thank Tanya Simms for her valuable comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Lacau, H., Gayden, T., Regueiro, M. et al. Afghanistan from a Y-chromosome perspective. Eur J Hum Genet 20, 1063–1070 (2012). https://doi.org/10.1038/ejhg.2012.59
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2012.59
Keywords
This article is cited by
-
Contrasting maternal and paternal genetic histories among five ethnic groups from Khyber Pakhtunkhwa, Pakistan
Scientific Reports (2022)
-
Mitochondrial DNA diversity in the Khattak and Kheshgi of the Peshawar Valley, Pakistan
Genetica (2020)
-
Afghanistan: conduits of human migrations identified using AmpFlSTR markers
International Journal of Legal Medicine (2019)
-
Dispersals of the Siberian Y-chromosome haplogroup Q in Eurasia
Molecular Genetics and Genomics (2018)
-
Phylogeography of human Y-chromosome haplogroup Q3-L275 from an academic/citizen science collaboration
BMC Evolutionary Biology (2017)