Abstract
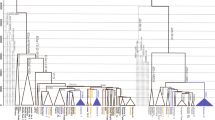
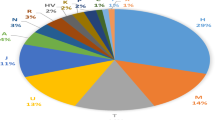
Haplogroup G, together with J2 clades, has been associated with the spread of agriculture, especially in the European context. However, interpretations based on simple haplogroup frequency clines do not recognize underlying patterns of genetic diversification. Although progress has been recently made in resolving the haplogroup G phylogeny, a comprehensive survey of the geographic distribution patterns of the significant sub-clades of this haplogroup has not been conducted yet. Here we present the haplogroup frequency distribution and STR variation of 16 informative G sub-clades by evaluating 1472 haplogroup G chromosomes belonging to 98 populations ranging from Europe to Pakistan. Although no basal G-M201* chromosomes were detected in our data set, the homeland of this haplogroup has been estimated to be somewhere nearby eastern Anatolia, Armenia or western Iran, the only areas characterized by the co-presence of deep basal branches as well as the occurrence of high sub-haplogroup diversity. The P303 SNP defines the most frequent and widespread G sub-haplogroup. However, its sub-clades have more localized distribution with the U1-defined branch largely restricted to Near/Middle Eastern and the Caucasus, whereas L497 lineages essentially occur in Europe where they likely originated. In contrast, the only U1 representative in Europe is the G-M527 lineage whose distribution pattern is consistent with regions of Greek colonization. No clinal patterns were detected suggesting that the distributions are rather indicative of isolation by distance and demographic complexities.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Yunusbayev B, Metspalu M, Järve M et al. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol Biol Evol 2011; 29: 359–365.
Balanovsky O, Dibirova K, Dybo A et al. Parallel evolution of genes and languages in the Caucasus region. Mol Biol Evol 2011; 28: 2905–2920.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ : Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered 2006; 61: 132–143.
Semino O, Passarino G, Oefner PJ et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 2000; 290: 1155–1159.
Nasidze I, Quinque D, Dupanloup I et al. Genetic evidence concerning the origins of South and North Ossetians. Ann Hum Genet 2004; 68: 588–599.
Goncalves R, Freitas A, Branco M et al. Y-chromosome lineages from Portugal, Madeira and Acores record elements of Sephardim and Berber ancestry. Ann Hum Genet 2005; 69: 443–454.
Capelli C, Brisighelli F, Scarnicci F et al. Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic-Neolithic encounter. Mol Phylogenet Evol 2007; 44: 228–239.
Battaglia V, Fornarino S, Al-Zahery N et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur J Hum Genet 2009; 17: 820–830.
King RJ, Ozcan SS, Carter T et al. Differential Y-chromosome Anatolian influences on the Greek and Cretan Neolithic. Ann Hum Genet 2008; 72: 205–214.
Cinnioglu C, King R, Kivisild T et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Hammer MF, Behar DM, Karafet TM et al. Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood. Hum Genet 2009; 126: 707–717.
Sengupta S, Zhivotovsky LA, King R et al. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 2006; 78: 202–221.
Semino O, Magri C, Benuzzi G et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004; 74: 1023–1034.
Zalloua PA, Xue Y, Khalife J et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet 2008; 82: 873–882.
King RJ, DiCristofaro J, Kouvatsi A et al. The coming of the Greeks to Provence and Corsica: Y-chromosome models of archaic Greek colonization of the western Mediterranean. BMC Evol Biol 2011; 11: 69.
Sims LM, Garvey D, Ballantyne J : Improved resolution haplogroup G phylogeny in the Y chromosome, revealed by a set of newly characterized SNPs. PLoS One 2009; 4: e5792.
Balanovsky O, Rootsi S, Pshenichnov A et al. Two sources of the Russian patrilineal heritage in their Eurasian context. Am J Hum Genet 2008; 82: 236–250.
Flores C, Maca-Meyer N, Gonzalez AM et al. Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography. Eur J Hum Genet 2004; 12: 855–863.
Barac L, Pericic M, Klaric IM et al. Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet 2003; 11: 535–542.
Pericic M, Lauc LB, Klaric IM, Janicijevic B, Rudan P : Review of croatian genetic heritage as revealed by mitochondrial DNA and Y chromosomal lineages. Croat Med J 2005; 46: 502–513.
Martinez L, Underhill PA, Zhivotovsky LA et al. Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau. Eur J Hum Genet 2007; 15: 485–493.
Luis JR, Rowold DJ, Regueiro M et al. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 2004; 74: 788–788.
Behar DM, Yunusbayev B, Metspalu M et al. The genome-wide structure of the Jewish people. Nature 2010; 466: 238–242.
Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ : Y-chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet 2008; 16: 374–386.
Chiaroni J, King RJ, Myres NM et al. The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations. Eur J Hum Genet 2010; 18: 348–353.
Kivisild T, Rootsi S, Metspalu M et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 2003; 72: 313–332.
Keller A, Graefen A, Ball M et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun 2012; 3.
de Knijff P, Kayser M, Caglia A et al. Chromosome Y microsatellites: population genetic and evolutionary aspects. Int J Legal Med 1997; 110: 134–149.
Kayser M, Caglia A, Corach D et al. Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 1997; 110: 141–149.
White PS, Tatum OL, Deaven LL, Longmire JL : New, male-specific microsatellite markers from the human Y chromosome. Genomics 1999; 57: 433–437.
Zhivotovsky LA, Underhill PA, Cinnioglu C et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 2004; 74: 50–61.
Zhivotovsky LA, Underhill PA, Feldman MW : Difference between evolutionarily effective and germ line mutation rate due to stochastically varying haplogroup size. Mol Biol Evol 2006; 23: 2268–2270.
Pichler I, Fuchsberger C, Platzer C et al. Drawing the history of the Hutterite population on a genetic landscape: inference from Y-chromosome and mtDNA genotypes. Eur J Hum Genet 2010; 18: 463–470.
Dulik MC, Zhadanov SI, Osipova LP et al. Mitochondrial DNA and Y Chromosome Variation Provides Evidence for a Recent Common Ancestry between Native Americans and Indigenous Altaians. Am J Hum Genet 2012; 90: 573.
Capelli C, Brisighelli F, Scarnicci F, Blanco-Verea A, Brion M, Pascali VL : Phylogenetic evidence for multiple independent duplication events at the DYS19 locus. Forensic Sci Int-Gen 2007; 1: 287–290.
Nei M : Molecular Evolutionary Genetics. New York: Columbia University Press, 1987.
Dulik MC, Osipova LP, Schurr TG : Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongolian expansions. PLoS One 2011; 6: e17548.
Haak W, Balanovsky O, Sanchez JJ et al. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol 2010; 8: e1000536.
Kharkov VN, Stepanov VA, Borinskaya SA et al. Gene pool structure of Eastern Ukrainians as inferred from the Y-chromosome haplogroups. Russ J Genet 2004; 40: 326–331.
Cavalli-Sforza L, Menozzi P, Piazza A : The History and Geography of Human Genes. Princeton: Princeton University Press, 1994.
Kaniewski D, Van Campo E, Van Lerberghe K et al. The Sea Peoples, from cuneiform tablets to carbon dating. PLoS One 2011; 6: e20232.
Achilli A, Olivieri A, Pala M et al. Mitochondrial DNA variation of modern Tuscans supports the near eastern origin of Etruscans. Am J Hum Genet 2007; 80: 759–768.
Vernesi C, Caramelli D, Dupanloup I et al. The Etruscans: a population-genetic study. Am J Hum Genet 2004; 74: 694–704.
Lacan M, Keyser C, Ricaut FX et al. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc Natl Acad Sci USA 2011; 108: 9788–9791.
Lacan M, Keyser C, Ricaut FX et al. Ancient DNA suggests the leading role played by men in the Neolithic dissemination. Proc Natl Acad Sci USA 2011; 108: 18255–18259.
Rosser ZH, Zerjal T, Hurles ME et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 2000; 67: 1526–1543.
Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA : Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet 2002; 70: 265–268.
Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J : High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet 2001; 68: 1019–1029.
Acknowledgements
JD and JC were supported by ANR program AFGHAPOP No BLAN07-9_222301’. RV and DMB thank the European Commission, Directorate-General for Research for FP7 Ecogene grant 205419. RV thanks the European Union Regional Development Fund for support through the Centre of Excellence in Genomics, the Estonian Ministry of Education and Research for the Basic Research grant SF 0270177As08. SR thanks the Estonian Science Foundation for grant 7445 and M Metspalu for grant 8973. EKK thanks the Russian Academy of Sciences Program for Fundamental Research ‘Biodiversity and dynamics of gene pools’, the Ministry of Education and Science of the Russian Federation for state contracts P-325 and 02.740.11.07.01, and the Russian Foundation for Basic Research for grants 04-04-48678-а and 07-04-01016-а. IK thanks the Russian Foundation for Basic Research for grant 08-06-97011 and the Grant of the President of the Russian Federation of state support for young Russian scientists MK-488.2006.4. OS thanks the Italian Ministry of the University: Progetti Ricerca Interesse Nazionale 2009 and FIRB-Futuro in Ricerca 2008 and Fondazione Alma Mater Ticinensins. AAL thanks the Sorenson Molecular Genealogy Foundation. PAU thanks Professor Carlos D Bustamante. MH and MHS are thankful to the ‘National Institute for Genetic Engineering and Biotechnology’, Tehran, Iran, and the ‘National Research Institute for Science policy’, Tehran, Iran, for providing the samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Rootsi, S., Myres, N., Lin, A. et al. Distinguishing the co-ancestries of haplogroup G Y-chromosomes in the populations of Europe and the Caucasus. Eur J Hum Genet 20, 1275–1282 (2012). https://doi.org/10.1038/ejhg.2012.86
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2012.86
Keywords
This article is cited by
-
Population genetic study of 17 Y-STR Loci of the Sorani Kurds in the Province of Sulaymaniyah, Iraq
BMC Genomics (2022)
-
The Dutch Y-chromosomal landscape
European Journal of Human Genetics (2020)
-
Sex-biased patterns shaped the genetic history of Roma
Scientific Reports (2020)
-
Middle eastern genetic legacy in the paternal and maternal gene pools of Chuetas
Scientific Reports (2020)
-
Ethnic Differences in the Risk Factors and Severity of Coronary Artery Disease: a Patient-Based Study in Iran
Journal of Racial and Ethnic Health Disparities (2018)