Abstract
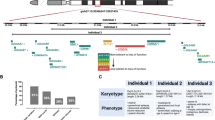
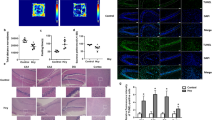
We identified a novel homozygous 15q13.3 microdeletion in a young boy, with a complex neurodevelopmental disorder characterized by severe cerebral visual impairment with additional signs of congenital stationary night blindness, congenital hypotonia with areflexia, profound intellectual disability, and refractory epilepsy. The mechanisms by which the genes in the deleted region exert their effect are unclear. In this paper, we probed the role of downstream effects of the deletions as a contributing mechanism to the molecular basis of the observed phenotype. We analyzed gene expression of lymphoblastoid cells derived from peripheral blood of the proband and his relatives to ascertain the relative effects of the homozygous and heterozygous deletions. We identified 267 genes with apparent differential expression between the proband with the homozygous deletion and 3 age- and sex-matched typically developing controls. Several of the differentially expressed genes are known to influence neurodevelopment and muscular function, and thus may contribute to the observed cognitive impairment and hypotonia. We further investigated the role of CHRNA7 by measuring TNFα modulation (a potentially important pathway in regulating synaptic plasticity). We found that the cell line with the homozygous deletion lost the ability to inhibit the activation of tumor necrosis factor-α secretion. Our findings suggest downstream genes that may have been altered by the 15q13.3 homozygous deletion, and thus contributed to the severe developmental encephalopathy of the proband. Furthermore, we show that a potentially important pathway in learning and development is affected by the deletion of CHRNA7.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Mulley JC, Dibbens LM : Chipping away at the common epilepsies with complex genetics: the 15q13.3 microdeletion shows the way. Genome Med 2009; 1: 33.
Ben-Shachar S, Lanpher B, German JR et al: Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet 2009; 46: 382–388.
Pagnamenta AT, Wing K, Akha ES et al: A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet 2009; 17: 687–692.
Miller DT, Shen Y, Weiss LA et al: Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet 2009; 46: 242–248.
Sharp AJ, Mefford HC, Li K et al: A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 2008; 40: 322–328.
Helbig I, Mefford HC, Sharp AJ et al: 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet 2009; 41: 160–162.
van Bon BW, Mefford HC, Menten B et al: Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet 2009; 46: 511–523.
Shinawi M, Schaaf CP, Bhatt SS et al: A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet 2009; 41: 1269–1271.
Gault J, Robinson M, Berger R et al: Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7). Genomics 1998; 52: 173–185.
Elmslie FV, Rees M, Williamson MP et al: Genetic mapping of a major susceptibility locus for juvenile myoclonic epilepsy on chromosome 15q. Hum Mol Genet 1997; 6: 1329–1334.
Neubauer BA, Fiedler B, Himmelein B et al: Centrotemporal spikes in families with rolandic epilepsy: linkage to chromosome 15q14. Neurology 1998; 51: 1608–1612.
Martin LF, Leonard S, Hall MH, Tregellas JR, Freedman R, Olincy A : Sensory gating and alpha-7 nicotinic receptor gene allelic variants in schizoaffective disorder, bipolar type. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 611–614.
Hoppman-Chaney N, Wain K, Seger PR, Superneau DW, Hodge JC : Identification of single gene deletions at 15q13.3: further evidence that CHRNA7 causes the 15q13.3 microdeletion syndrome phenotype. Clin Genet 2012, e-pub ahead of print 9 July 2012.
Masurel-Paulet A, Andrieux J, Callier P et al: Delineation of 15q13.3 microdeletions. Clin Genet 2010; 78: 149–161.
LePichon JB, Bittel DC, Graf WD, Yu S : A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7, and other homozygously deleted genes. Am J Med Genet A 2010; 152A: 1300–1304.
Endris V, Hackmann K, Neuhann TM et al: Homozygous loss of CHRNA7 on chromosome 15q13.3 causes severe encephalopathy with seizures and hypotonia. Am J Med Genet A 2010; 152A: 2908–2911.
Liao J, DeWard SJ, Madan-Khetarpal S, Surti U, Hu J : A small homozygous microdeletion of 15q13.3 including the CHRNA7 gene in a girl with a spectrum of severe neurodevelopmental features. Am J Med Genet A 2011; 155A: 2795–2800.
Audo I, Kohl S, Leroy BP et al: TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet 2009; 85: 720–729.
Li Z, Sergouniotis PI, Michaelides M et al: Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet 2009; 85: 711–719.
Nakamura M, Sanuki R, Yasuma TR et al: TRPM1 mutations are associated with the complete form of congenital stationary night blindness. Mol Vis 2010; 16: 425–437.
van Genderen MM, Bijveld MM, Claassen YB et al: Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet 2009; 85: 730–736.
Webber C : Functional enrichment analysis with structural variants: pitfalls and strategies. Cytogenet Genome Res 2011; 135: 277–285.
Stellwagen D, Malenka RC : Synaptic scaling mediated by glial TNF-alpha. Nature 2006; 440: 1054–1059.
Bittel DC, Kibiryeva N, Sell SM, Strong TV, Butler MG : Whole genome microarray analysis of gene expression in Prader-Willi syndrome. Am J Med Genet A 2007; 143: 430–442.
Bittel DC, Kibiryeva N, McNulty SG, Driscoll DJ, Butler MG, White RA : Whole genome microarray analysis of gene expression in an imprinting center deletion mouse model of Prader-Willi syndrome. Am J Med Genet A 2007; 143: 422–429.
Eisenhart C : The assumptions underlying the analysis of variance. Biometrics 1947; 3: 1–21.
Wang H, Yu M, Ochani M et al: Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature 2003; 421: 384–388.
Matsukawa A : STAT proteins in innate immunity during sepsis: lessons from gene knockout mice. Acta Med Okayama 2007; 61: 239–245.
Lee ST, Chu K, Sinn DI et al: Erythropoietin reduces perihematomal inflammation and cell death with eNOS and STAT3 activations in experimental intracerebral hemorrhage. J Neurochem 2006; 96: 1728–1739.
Chakravarti S, Magnuson T : Localization of mouse lumican (keratan sulfate proteoglycan) to distal chromosome 10. Mamm Genome 1995; 6: 367–368.
Yeh LK, Liu CY, Kao WW et al: Knockdown of zebrafish lumican gene (zlum) causes scleral thinning and increased size of scleral coats. J Biol Chem 2010; 285: 28141–28155.
Shen JX, Yakel JL : Nicotinic acetylcholine receptor-mediated calcium signaling in the nervous system. Acta Pharmacol Sin 2009; 30: 673–680.
Russo P, Taly A : α7-Nicotinic acetylcholine receptors: an old actor for new different roles. Curr Drug Targets 2012; 13: 574–578.
Rosas-Ballina M, Olofsson PS, Ochani M et al: Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science 2011; 334: 98–101.
Shen JX, Yakel JL : Functional alpha7 nicotinic ACh receptors on astrocytes in rat hippocampal CA1 slices. J Mol Neurosci 2012; 48: 14–21.
Diefenbach A, Schindler H, Rollinghoff M, Yokoyama WM, Bogdan C : Requirement for type 2 NO synthase for IL-12 signaling in innate immunity. Science 1999; 284: 951–955.
Hong MS, Song JY, Yun DH, Cho JJ, Chung JH : Increase of NADPH-diaphorase expression in hypothalamus of stat4 knockout mice. Korean J Physiol Pharmacol 2009; 13: 337–341.
Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM : Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci 2007; 8: 766–775.
Dogra C, Srivastava DS, Kumar A : Protein-DNA array-based identification of transcription factor activities differentially regulated in skeletal muscle of normal and dystrophin-deficient mdx mice. Mol Cell Biochem 2008; 312: 17–24.
Ma Y, Liu H, Tu-Rapp H et al: Fas ligation on macrophages enhances IL-1R1-Toll-like receptor 4 signaling and promotes chronic inflammation. Nat Immunol 2004; 5: 380–387.
Choi C, Benveniste EN : Fas ligand/Fas system in the brain: regulator of immune and apoptotic responses. Brain Res Brain Res Rev 2004; 44: 65–81.
Bentires-Alj M, Kontaridis MI, Neel BG : Stops along the RAS pathway in human genetic disease. Nat Med 2006; 12: 283–285.
Newbern JM, Li X, Shoemaker SE et al: Specific functions for ERK/MAPK signaling during PNS development. Neuron 2011; 69: 91–105.
Girirajan S, Eichler EE : Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet 2010; 19: R176–R187.
Acknowledgements
We thank the patient and his family for their cooperation. This research was supported by a generous endowment from the State of Kansas Fraternal order of Eagles.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Le Pichon, JB., Yu, S., Kibiryeva, N. et al. Genome-wide gene expression in a patient with 15q13.3 homozygous microdeletion syndrome. Eur J Hum Genet 21, 1093–1099 (2013). https://doi.org/10.1038/ejhg.2013.1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.1
Keywords
This article is cited by
-
Local and global chromatin interactions are altered by large genomic deletions associated with human brain development
Nature Communications (2018)
-
Modulatory effects of α7 nAChRs on the immune system and its relevance for CNS disorders
Cellular and Molecular Life Sciences (2016)