Abstract
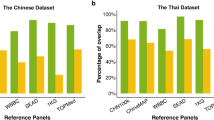
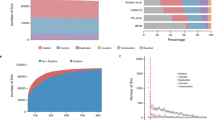
Population stratification acts as a confounding factor in genetic association studies and may lead to false-positive or false-negative results. Previous studies have analyzed the genetic substructures in Han Chinese population, the largest ethnic group in the world comprising ∼20% of the global human population. In this study, we examined 5540 Han Chinese individuals with about 1 million single-nucleotide polymorphisms (SNPs) and screened a panel of ancestry informative markers (AIMs) to facilitate the discerning and controlling of population structure in future association studies on Han Chinese. Based on genome-wide data, we first confirmed our previous observation of the north–south differentiation in Han Chinese population. Second, we developed a panel of 150 validated SNP AIMs to determine the northern or southern origin of each Han Chinese individual. We further evaluated the performance of our AIMs panel in association studies in simulation analysis. Our results showed that this AIMs panel had sufficient power to discern and control population stratification in Han Chinese, which could significantly reduce false-positive rates in both genome-wide association studies (GWAS) and candidate gene association studies (CGAS). We suggest this AIMs panel be genotyped and used to control and correct population stratification in the study design or data analysis of future association studies, especially in CGAS which is the most popular approach to validate previous reports on genetic associations of diseases in post-GWAS era.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Pritchard JK, Rosenberg NA : Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 1999; 65: 220–228.
Freedman ML, Reich D, Penney KL et al: Assessing the impact of population stratification on genetic association studies. Nat Genet 2004; 36: 388–393.
Marchini J, Cardon LR, Phillips MS, Donnelly P : The effects of human population structure on large genetic association studies. Nat Genet 2004; 36: 512–517.
Campbell CD, Ogburn EL, Lunetta KL et al: Demonstrating stratification in a European American population. Nat Genet 2005; 37: 868–872.
Price AL, Butler J, Patterson N et al: Discerning the ancestry of European Americans in genetic association studies. PLoS Genet 2008; 4: e236.
Tian C, Plenge RM, Ransom M et al: Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet 2008; 4: e4.
Paschou P, Drineas P, Lewis J et al: Tracing sub-structure in the European American population with PCA-informative markers. PLoS Genet 2008; 4: e1000114.
Xu S, Yin X, Li S et al: Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet 2009; 85: 762–774.
Chen J, Zheng H, Bei J-X et al: Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am J Hum Genet 2009; 85: 775–785.
Suo C, Xu H, Khor C-C et al: Natural positive selection and north-south genetic diversity in East Asia. Eur J Hum Genet 2012; 20: 102–110.
Abecasis GR, Auton A, Brooks LD et al: An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65.
Altshuler DM, Ra Gibbs, Peltonen L et al: Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–58.
Korn JM, Kuruvilla FG, McCarroll SA et al: Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008; 40: 1253–1260.
Tang H, Peng J, Wang P, Risch NJ : Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol 2005; 28: 289–301.
Patterson N, Price AL, Reich D : Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
Weir BS, Cockerham CC : Estimating F-statistics for the analysis of population structure. Evolution 1984; 38: 1358–1370.
Ding L, Wiener H, Abebe T et al: Comparison of measures of marker informativeness for ancestry and admixture mapping. BMC genomics 2011; 12: 622.
Rosenberg NA, Li LM, Ward R, Pritchard JK : Informativeness of genetic markers for inference of ancestry. Am J Hum Genet 2003; 73: 1402–1422.
Armitage P : Tests for linear trends in proportions and frequencies. Biometrics 1955; 11: 375–386.
Benjamini Y, Hochberg Y : Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Statist Soc B 1995; 57: 289–300.
Devlin B, Roeder K : Genomic control for association studies. Biometrics 1999; 55: 997–1004.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Su B, Xiao J, Underhill P et al: Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 1999; 65: 1718–1724.
Lao O, Lu TT, Nothnagel M et al: Correlation between genetic and geographic structure in Europe. Curr Biol 2008; 18: 1241–1248.
Qu HQ, Li Q, Xu S et al: Ancestry informative marker set for han chinese population. G3 2012; 2: 339–341.
Seldin MF, Price AL : Application of ancestry informative markers to association studies in European Americans. PLoS Genet 2008; 4: e5.
Acknowledgements
These studies were supported by the National Science Foundation of China (NSFC) grants (31171218; 30971577), by the Shanghai Rising-Star Program 11QA1407600, and by the Science Foundation of the Chinese Academy of Sciences (KSCX2-EW-Q-1-11; KSCX2-EW-R-01-05; KSCX2-EW-J-15-05). SX is Max Planck Independent Research Group Leader and member of CAS Youth Innovation Promotion Association. SX also gratefully acknowledges the support of the National Program for Top-notch Young Innovative Talents and the support of KCWong Education Foundation, Hong Kong. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Qin, P., Li, Z., Jin, W. et al. A panel of ancestry informative markers to estimate and correct potential effects of population stratification in Han Chinese. Eur J Hum Genet 22, 248–253 (2014). https://doi.org/10.1038/ejhg.2013.111
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.111
Keywords
This article is cited by
-
Ancestry informative markers (AIMs) for Korean and other East Asian and South East Asian populations
International Journal of Legal Medicine (2019)
-
Identifying novel microhaplotypes for ancestry inference
International Journal of Legal Medicine (2019)
-
Homozygous p.Ser267Phe in SLC10A1 is associated with a new type of hypercholanemia and implications for personalized medicine
Scientific Reports (2017)
-
A single-tube 27-plex SNP assay for estimating individual ancestry and admixture from three continents
International Journal of Legal Medicine (2016)
-
Selection of highly informative SNP markers for population affiliation of major US populations
International Journal of Legal Medicine (2016)